Help Page
Help section page of alfaNET: Here you will find a guide to navigate through the database. If you have any questions that are not covered on this page, please send an email to Raghav Kataria.
Introduction
Alfalfa is an extensively grown forage crop, and is widely adapted due to its high biomass, and role in soil-water conservation. In the past few years, Bacterial Stem Blight has emerged as a major threat for agriculture industry. This disease does not have a cure yet, and thus many efforts have been made to find a solution to this devastating condition. There are challenges in the generation of high-yield resistant cultivars, in part due to the limited and sparse knowledge about the mechanisms that are used by the Pseudomonas bacteria to proliferate the infection in alfalfa plants. This database has been implemented to provide the annotation of Pseudomonas proteome, as well as the host-pathogen interactomics tool, a platform to compare the predicted interactome of host-pathogen system.
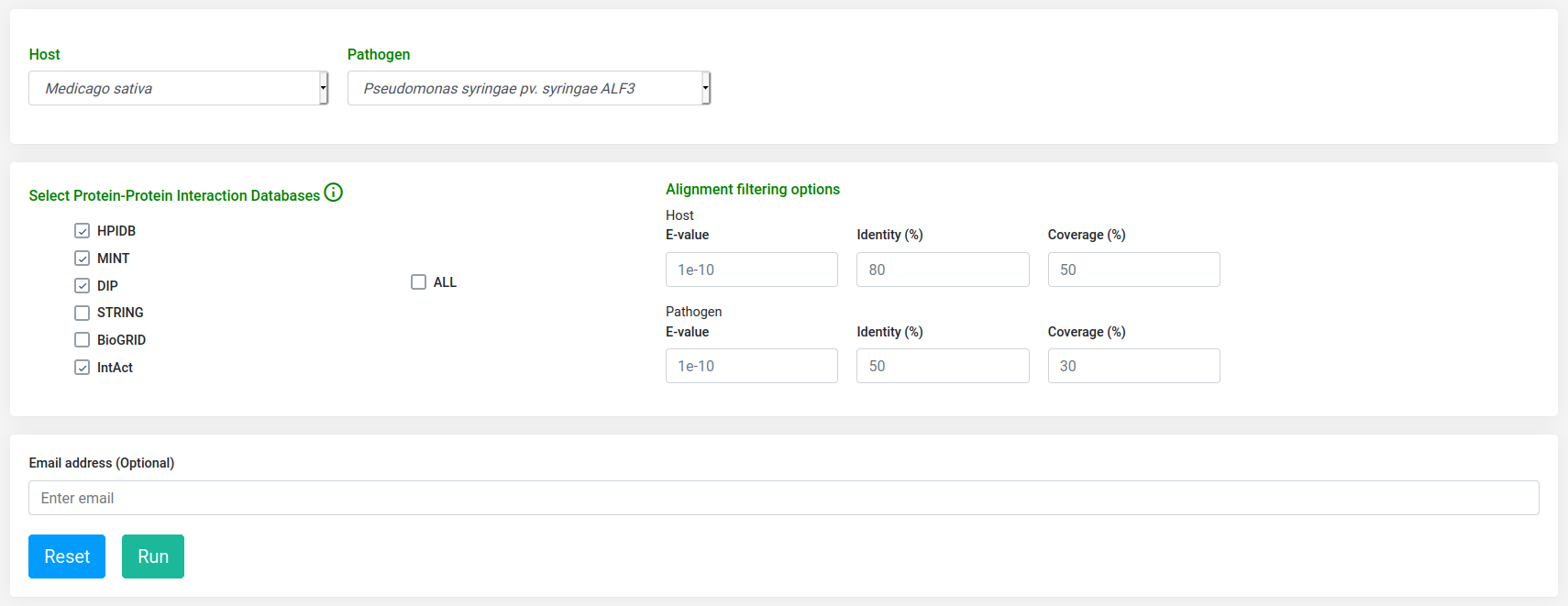
Host-pathogen interactomics module
With Interactomics tool, a user can find the interactions between host (Medicago sativa) and pathogen (Pseudomonas syringae ALF3). Other options for the user in this module include selecting which protein-protein database will be used as a template in the prediction process, or to define BLASTp alignment filters to determine homolog proteins.
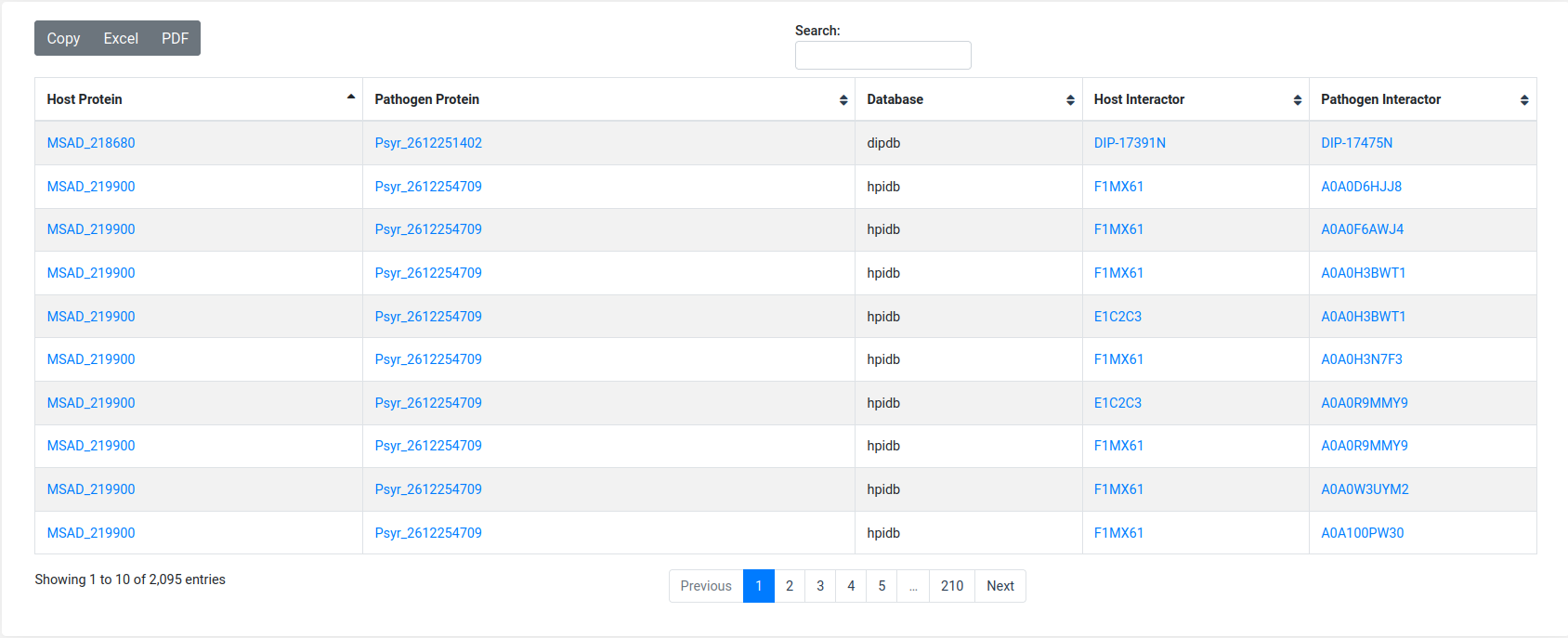
When a job is submitted it will be assigned to a unique identifier that user can access to check the status of the job (queried, running or done). After the job is completed it will display the results in an enriched table with the option to sort the content by column or to be filtered by keyword; result table can be downloaded in excel or pdf format, or copy as clipboard. Users also can provide an email address to receive a notification when the job completes.

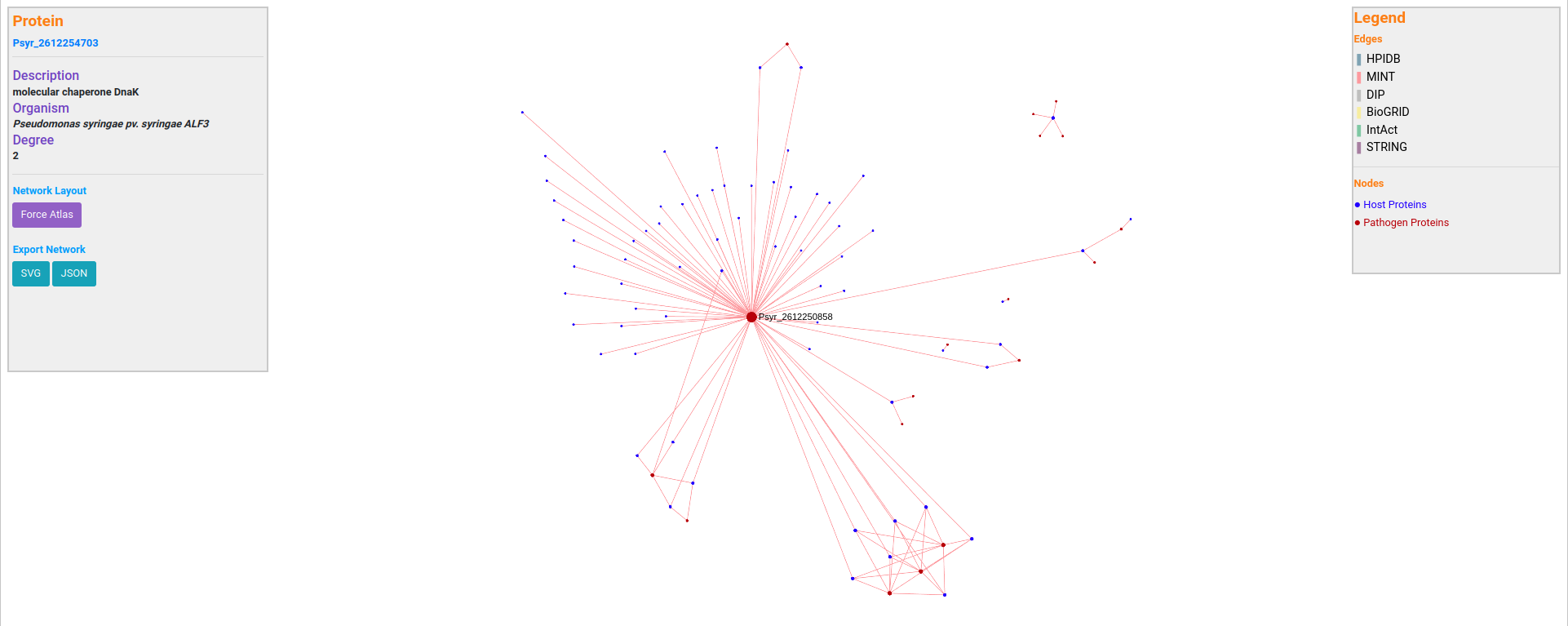
alfaNET provides a network visualization platform, implemented using SigmaJS. This plugin was specially chosen given its performance at displaying large networks. From the host-pathogen network visualization, a user can visualize a set of traits for each node (species, description, degree), and also can easily identify hub nodes (nodes with a higher number of edges). This is useful, as hub nodes have been found crucial in infectious disease pathways. A user is not limited to the network analysis that is provided through our database, the resulted network can be further examined in any network analyzer that could handle JSON or tabular network files.
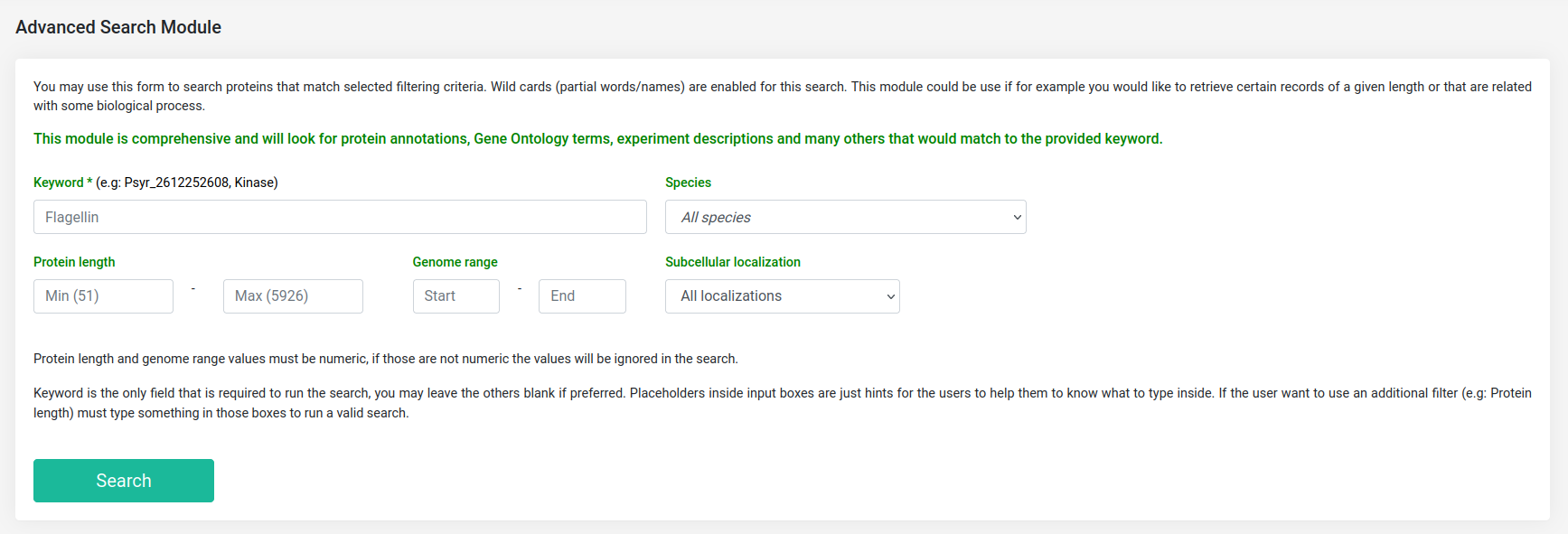
Advanced search module
This tool provides an advanced search module that can be used to search for proteins that fulfill a selected filtering criterion, means that for a given keyword plus a set of filtering parameters, this module will look up for any record that match. This search module is comprehensive and will look for protein annotations, GO terms, experiment descriptions and many others that would match to the provided keyword. Additionally, a basic option to perform a quick search of a protein accession is available at all the pages of alfaNET, and both the advanced and the basic search will display the complete information that can be obtained from our database records.
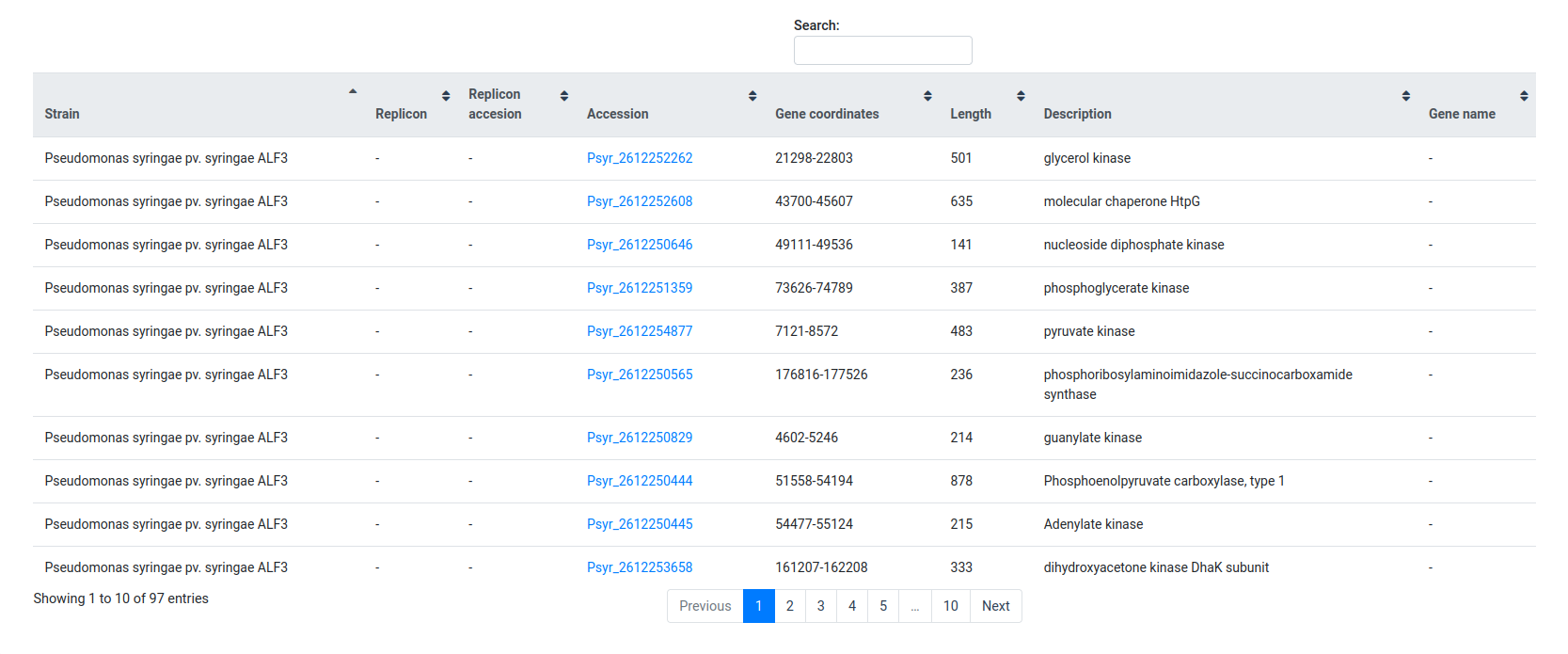
After the search is done, it will display a table with all the records that matched the selected filtering criterion.
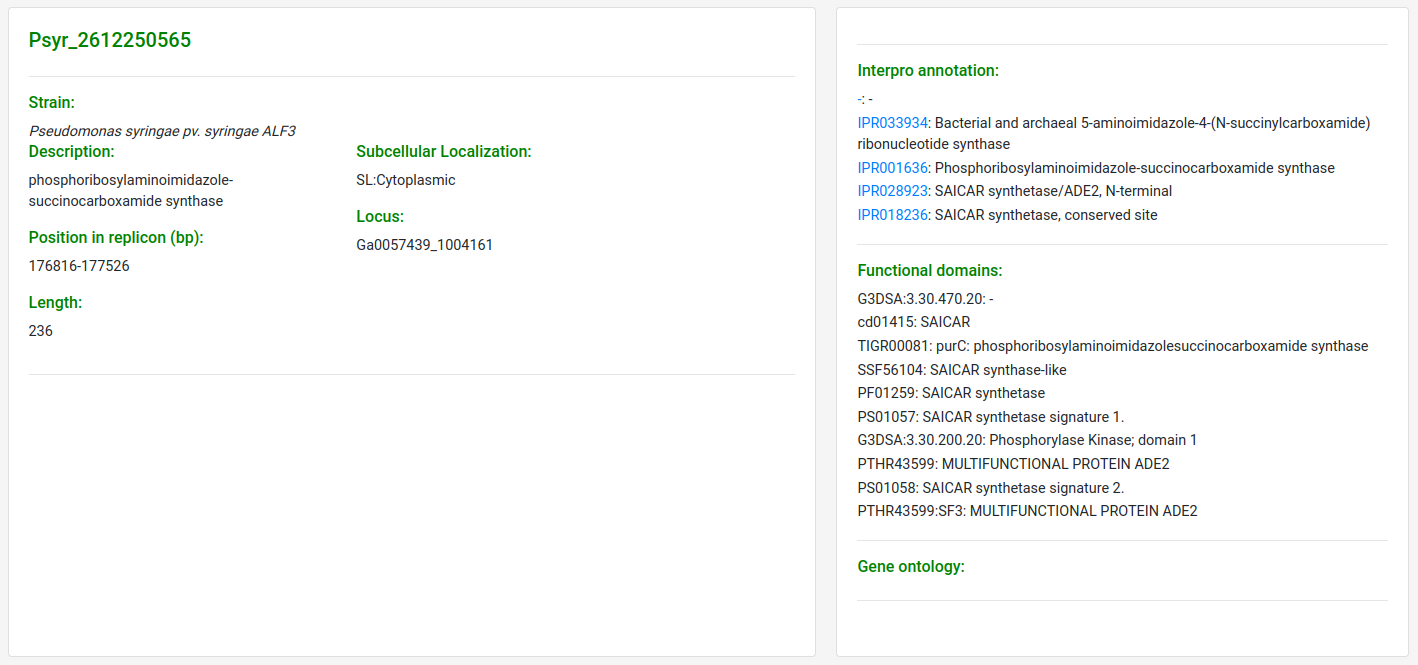
If click at one of the accessions links it will redirect the user to a new page in which is displayed the complete information that can be obtained from our database records, including functional domain annotation, GO terms, subcellular localization, protein length, locus, etc.
BLAST server
BLAST was implemented locally in our server to provide to the user the functionality of homology sequence search. In addition to the proteome datasets, a user can select to query its sequences against all the proteomes (default option). User can upload either a nucleotide or amino acid sequence, and the system will automatically detect the specific program (BLASTp or BLASTx) to be performed.
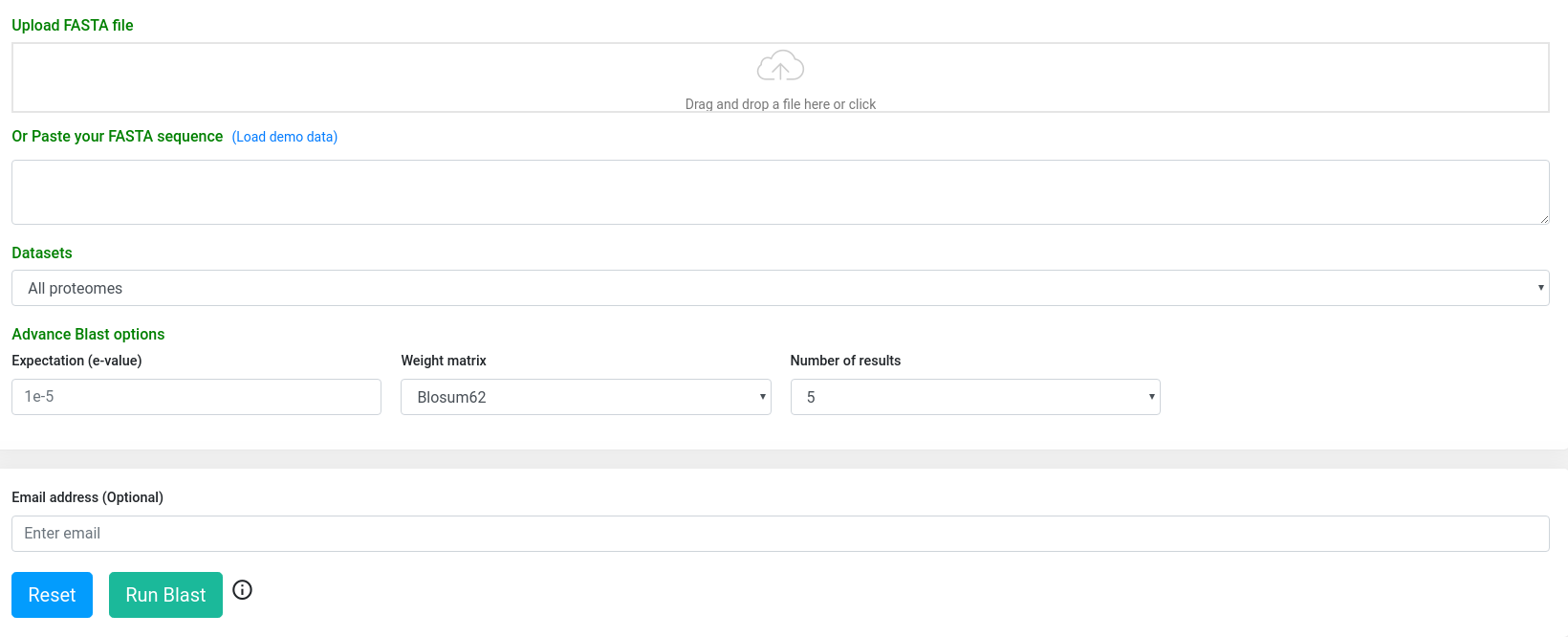
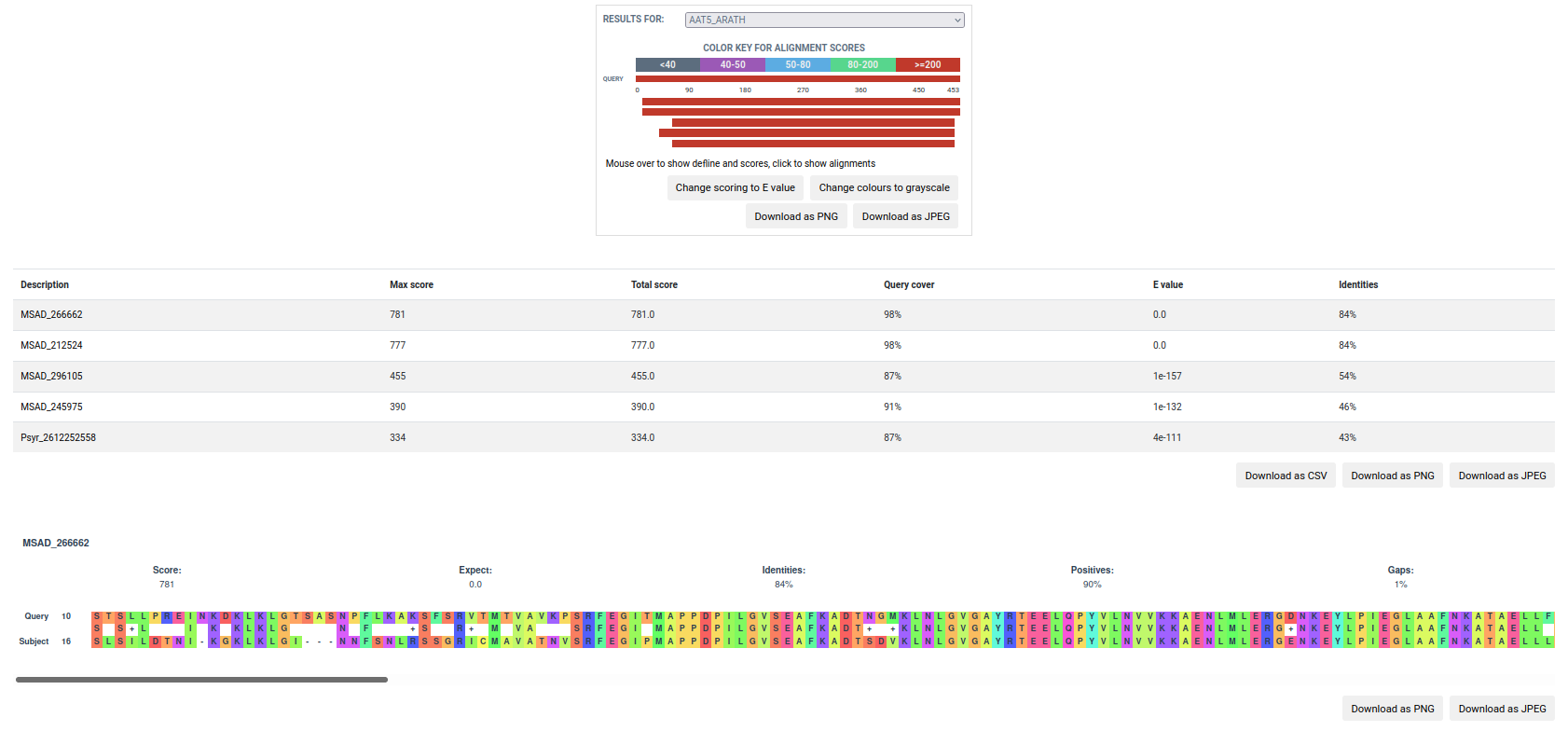
The BLAST result page provides a summarized version, whereby the user can download the alignments in tabular or standard alignment format. Also, there is a more detailed option in which the alignments are visualized in an enriched mode.
Summarized
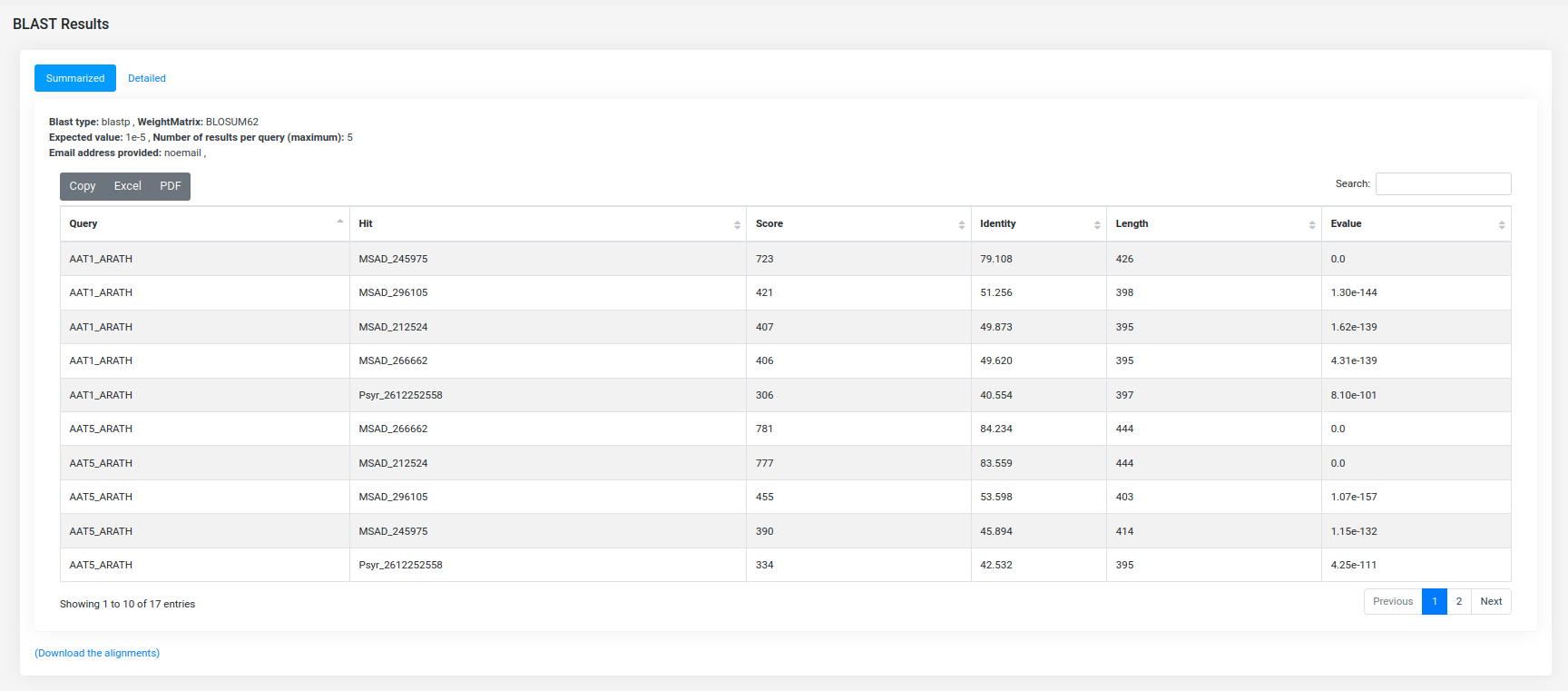
Detailed
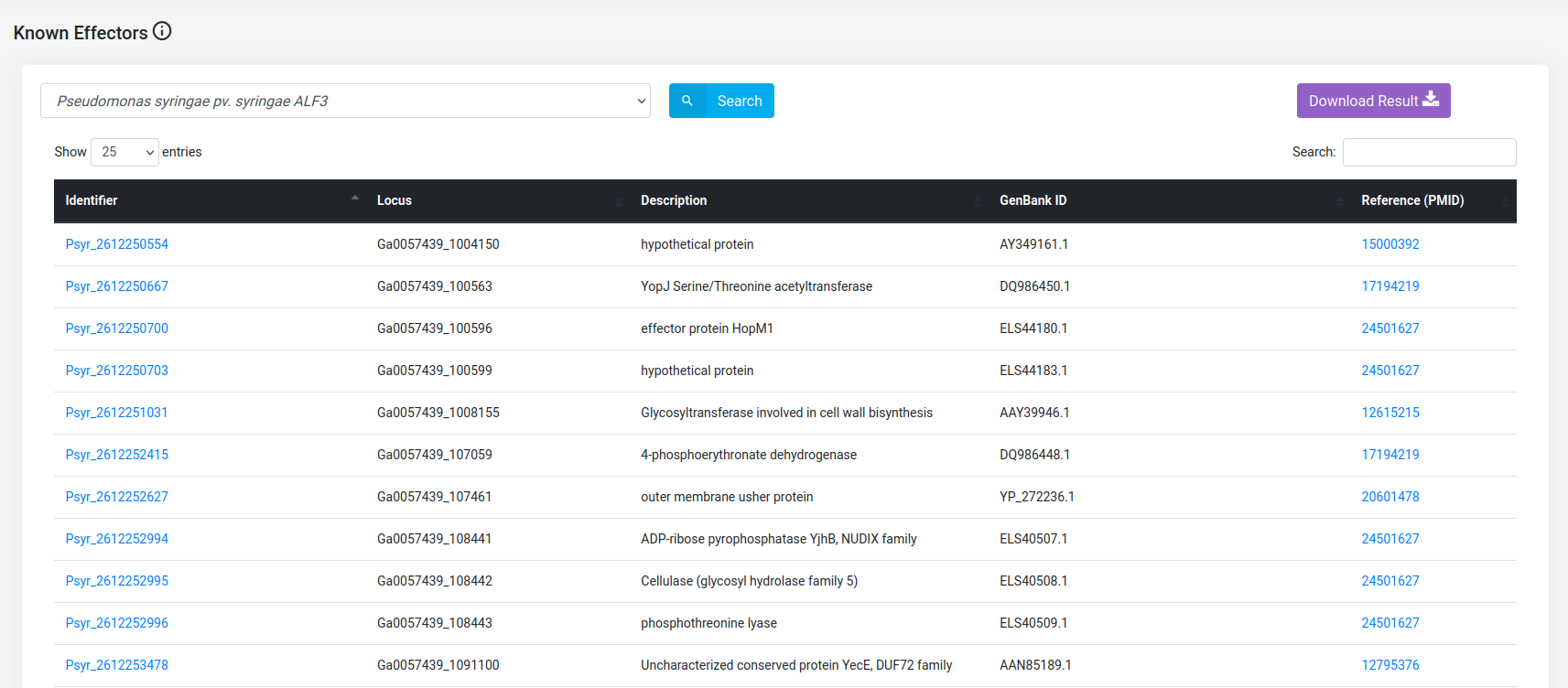
Features
Data collected from the literature or resulted from the annotation pipeline of alfaNET is presented in nine (9) search modules. In eight of those modules, Pseudomonas protein annotation can be retrieved (Protein Annotations, Subcellular Localization Annotation, Gene Ontology (GO) Term Annotation, Functional Domain Mappings (InterPro), Ice-nucleation proteins, Known effectors, Virulence effectors, and Predicted effectors (EffectiveDB)). In addition, the module "Host-pathogen interactions" is available to gather interactions concerning those Alfalfa proteins that were found related with Bacterial Stem Blight on the basis of interolog-based computational approach. The module data will be displayed according to the dataset selected.
Examples
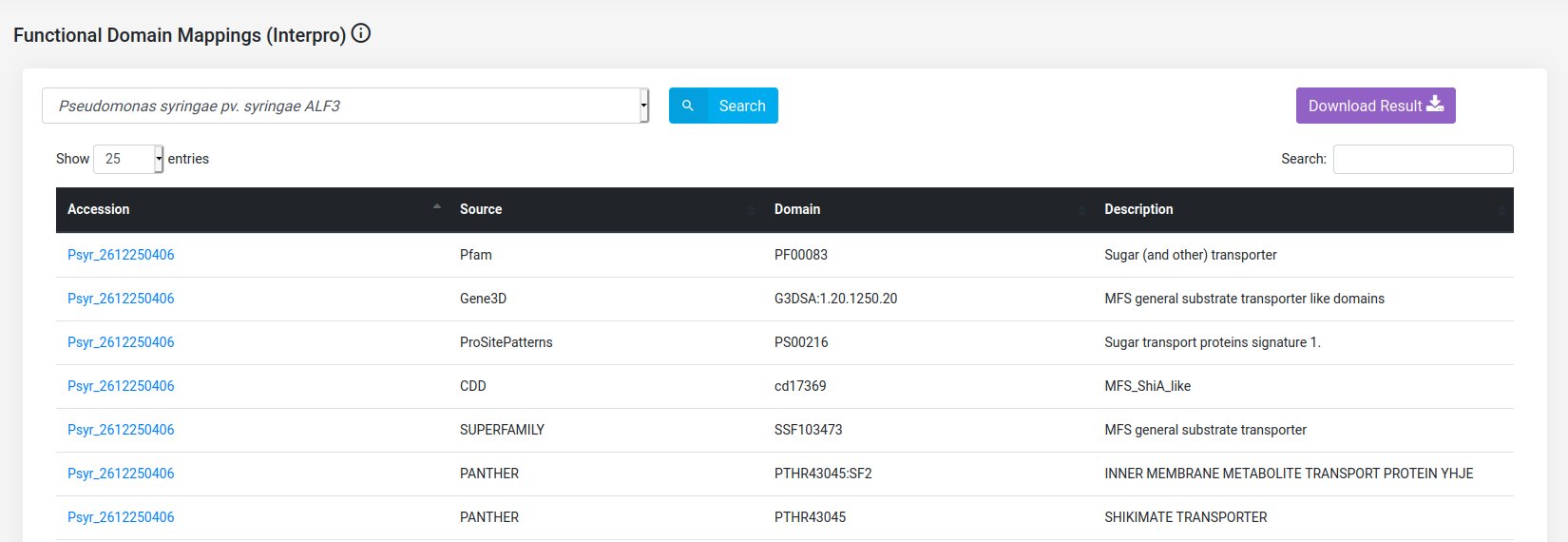
(a) Fetch the functional domains of Pseudomonas syringae pv. syringae ALF3 proteins.
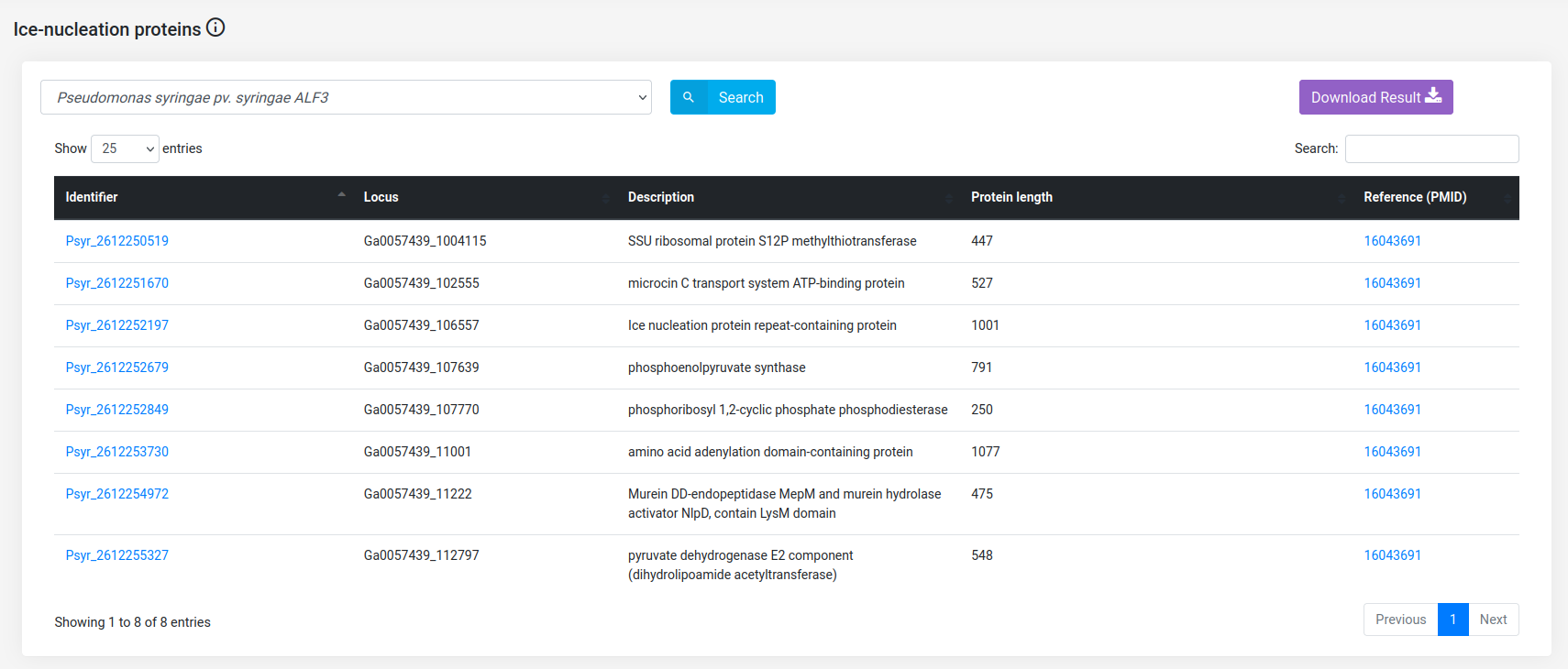
(b) Get the Ice-Nucleation Proteins (INPs).
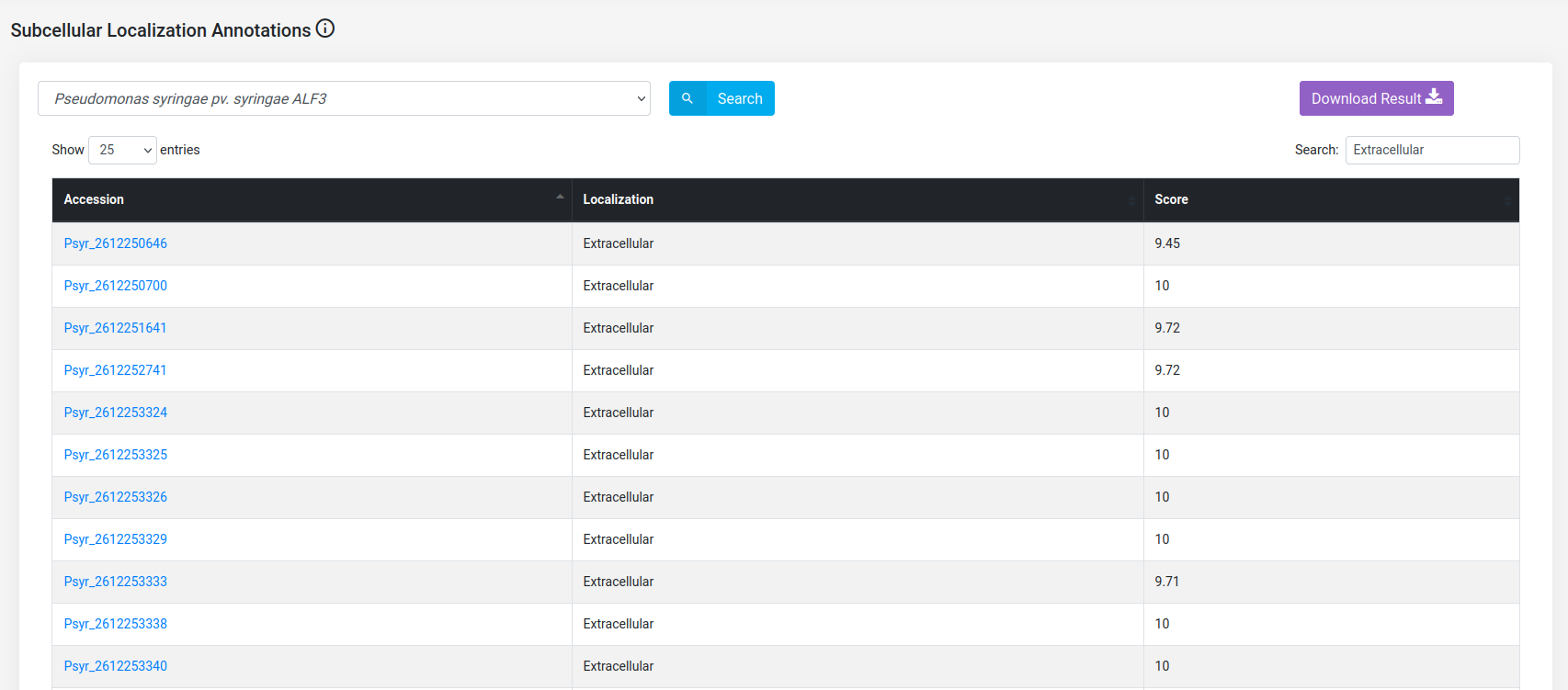
(c) Obtain the extracellular proteins of Pseudomonas syringae pv. syringae ALF3 proteins.
(d) Retrieve the known secreted effectors of Pseudomonas syringae pv. syringae ALF3.