How to use ProFeatX?
Introduction
This is a guide to help you use ProFeatX from the data input step to the results pageusing the demo data that is available for everyone, so that you can replicate the results. If you have any questions that are not covered in this page, send an email to david.guevara@usu.edu.
Data input
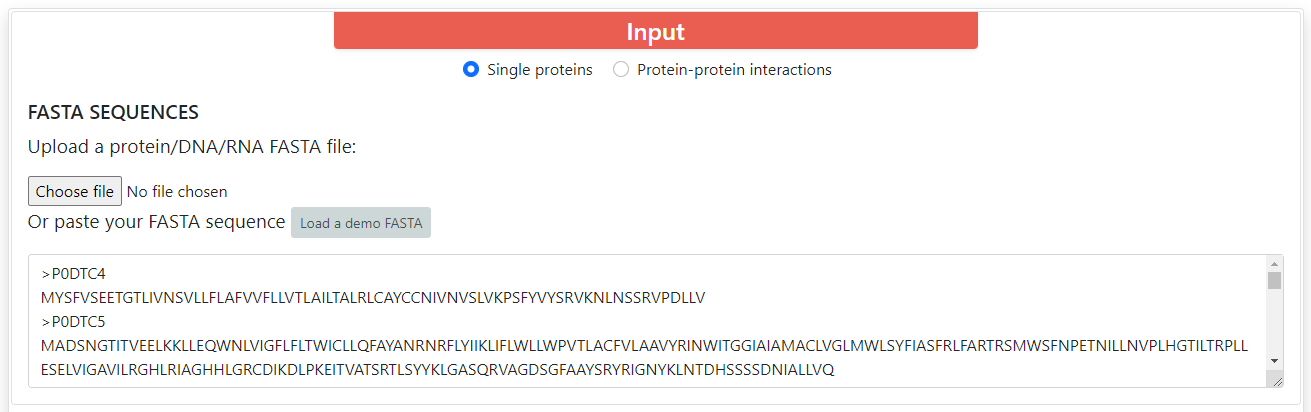
For all encodings the accepted input must be proteins or DNA/RNA sequences in FASTA format. You can upload a file or copy and paste a text for the sequences. You can find a button which says Load a demo FASTA, which will fill the input boxes with several protein sequences.
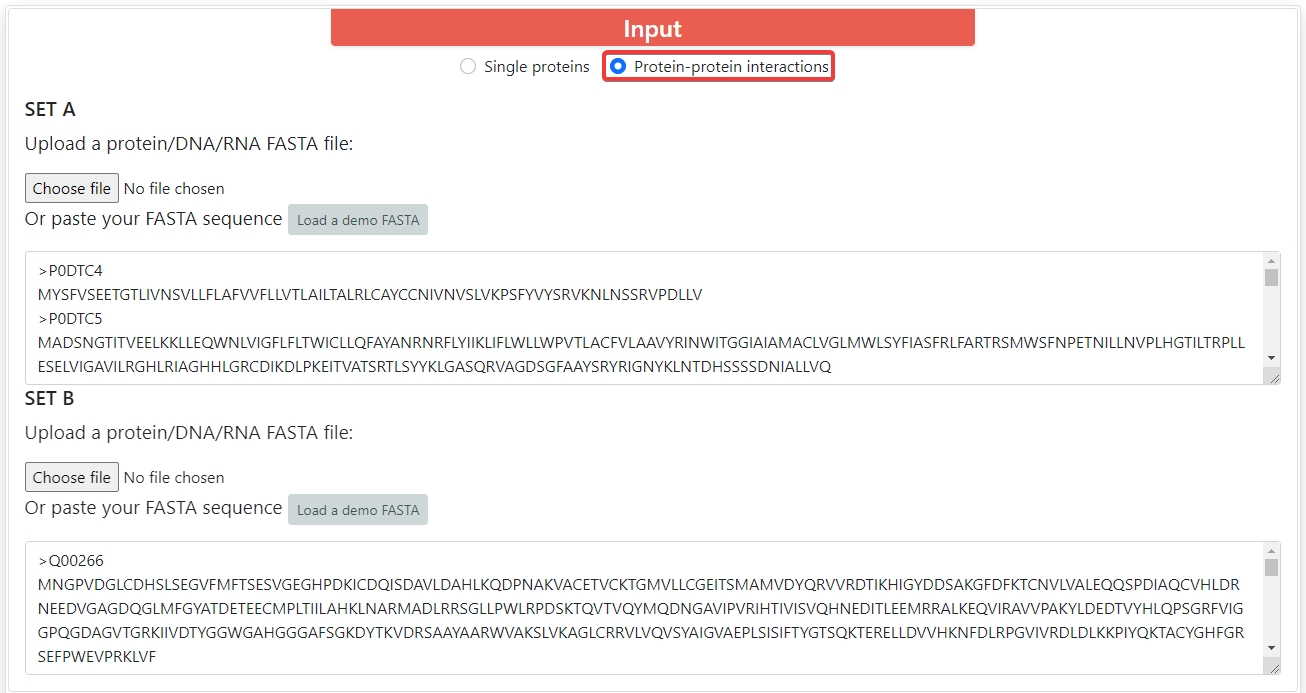
ProFeatX supports both single proteins by default and protein-protein interactions (PPI) encoding. To encode PPIs, you can click on Protein-protein interactions. You will be able to input two sets of proteins, and ProFeatX will join every sequence from set A with every sequence from set B, so if set A has 10 sequences, and set B has 5 sequences, the output will contain 50 encoded interactions.
WARNING: ProFeatX server supports up to 10,000 encoded sequences or interactions.
Encoding selection
ProFeatX server can encode your sequences in 32 different descriptors. These descriptors are organized into 10 groups.
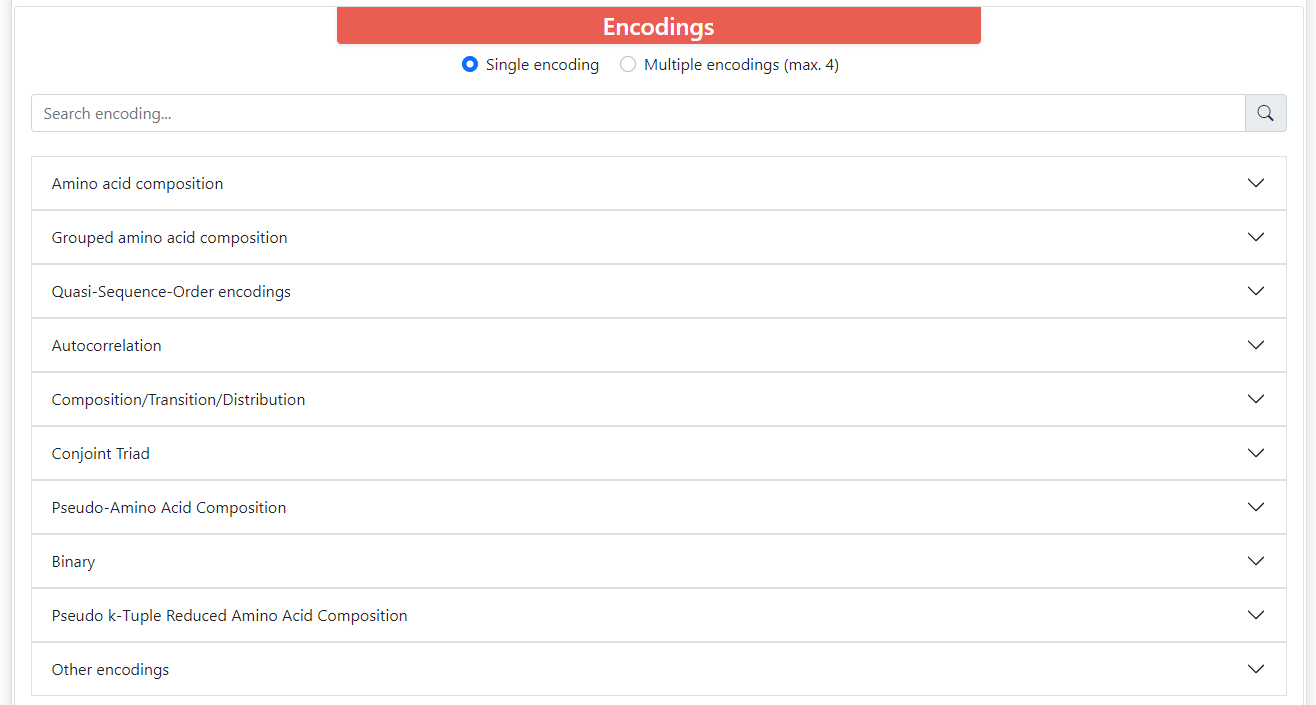
Above these groups there is a filter where you can type the name of the descriptors you want and it will filter the groups that contain descriptors with such name. All descriptors have its long name, short name, and vector size. Some of them depend on the length of the sequence, and these require that all the sequences within sets are the same length. Also, most encodings have parameters that you can modify.
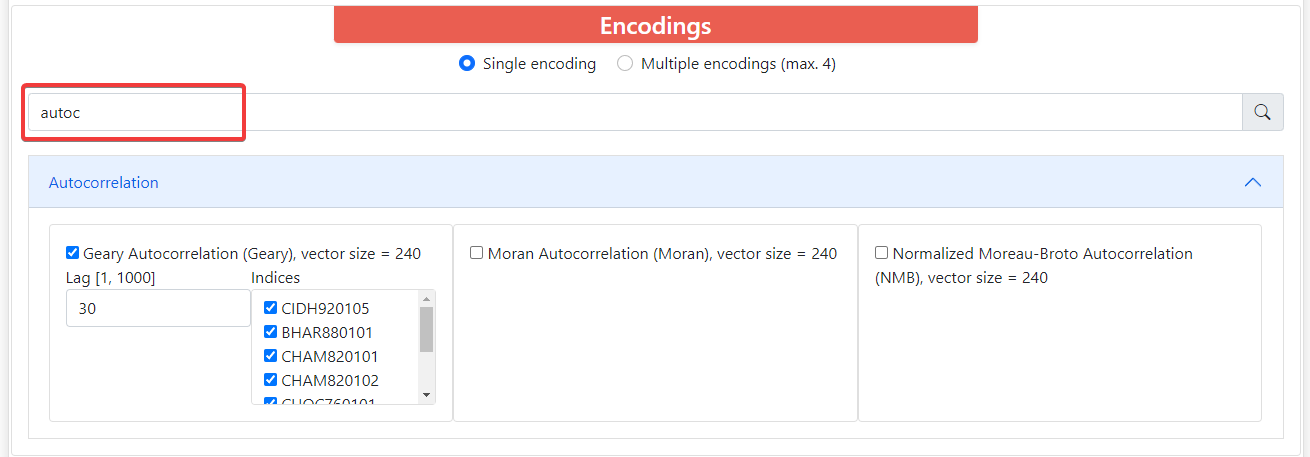
By default, ProFeatX only lets you select one encoding. However, you can also encode your sequences into up to 4 descriptors at the same time. To do this, click on Multiple encodings (max. 4) and it will let you choose 1-4 different encodings.
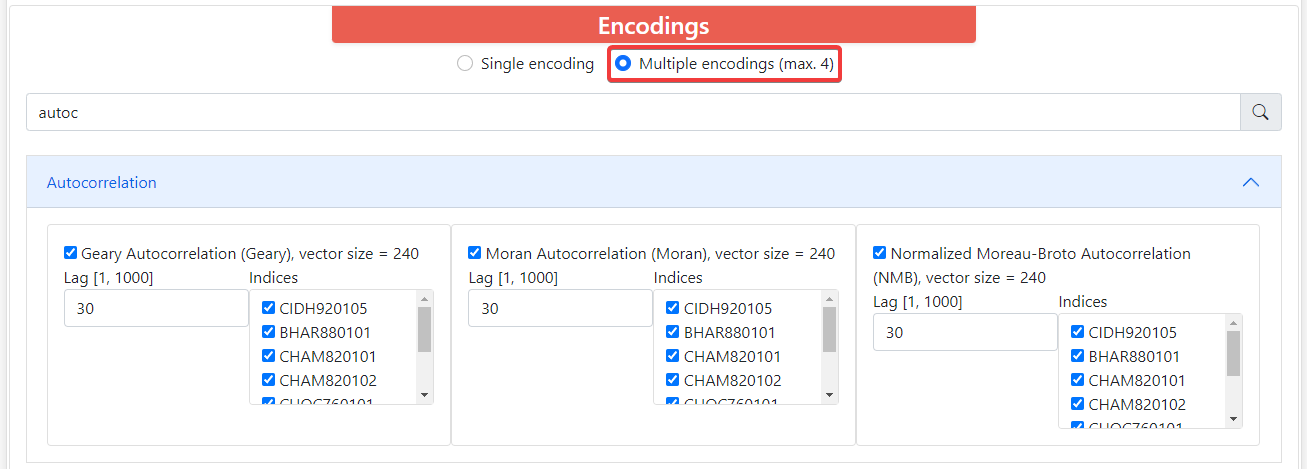
If you try to pick a fifth option, the webpage will not let you do it, and you will have to uncheck another box in order to choose a new fourth encoding.
Submit
The Submit section has a text box where you can type your email, so we tell you whenever your job is done running. Below you can find a button for resetting the form and the button for submitting your job. After submitting your job, if you stay in the same page, it will redirect you to the results page whenever it is done predicting.
Results
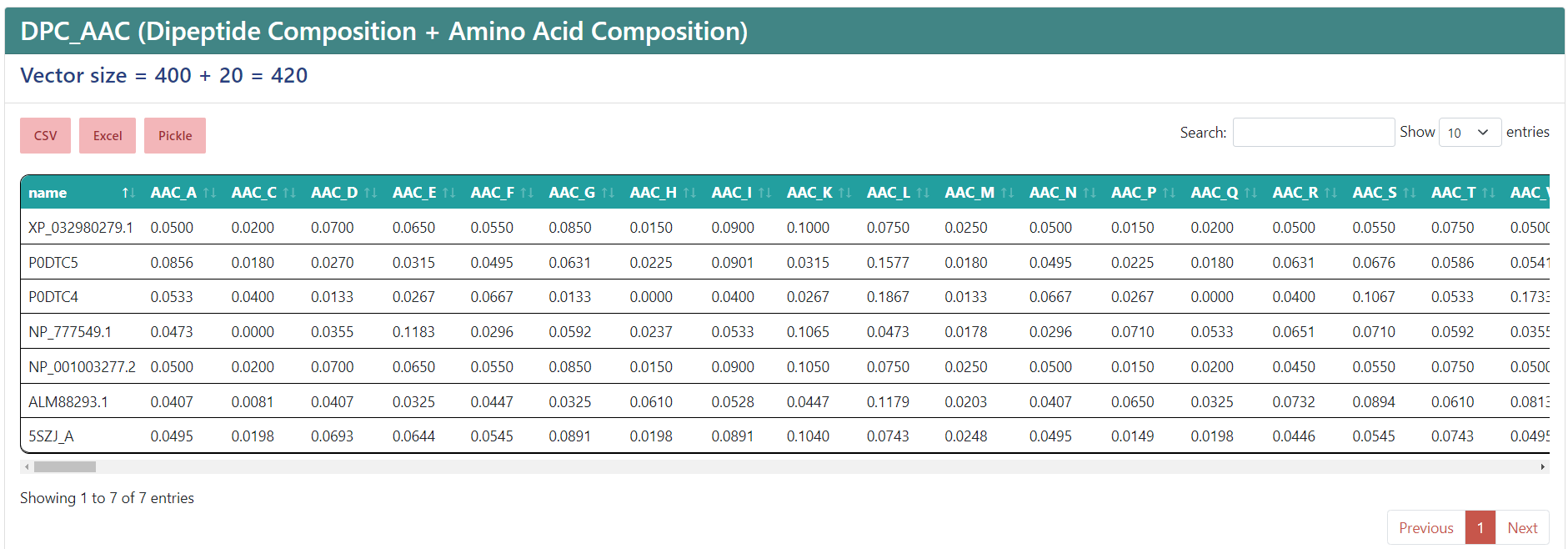
For the following examples we submitted our jobs filling them with the Load a demo FASTA button. At the top you will find the name of the selected encoding. If you selected multiple descriptors, they will appear separated by an underscore, like the example, where AAC and DPC were chosen, and the vector size, that is, the number of features. Underneath, you will find three buttons to export the result to a CSV, an Excel file or a Pickle (.pkl) file that contains a Pandas dataframe with the data. Also, to the right, there is a filter to search a sequence you want or a value you need, and a dropdown list to modify how many entries are shown in a single page.
The results table shows as many entries as sequences you input. The columns shown are the name of the sequence and the features, of which names come from the encoding the column corresponds to, an underscore, and a name associated to the value. If you only selected one descriptor, then only the associated value name will be shown, so instead of showing "AAC_A", it will show just "A".
WARNING: The displayed name of your sequences will be truncated after the first space.
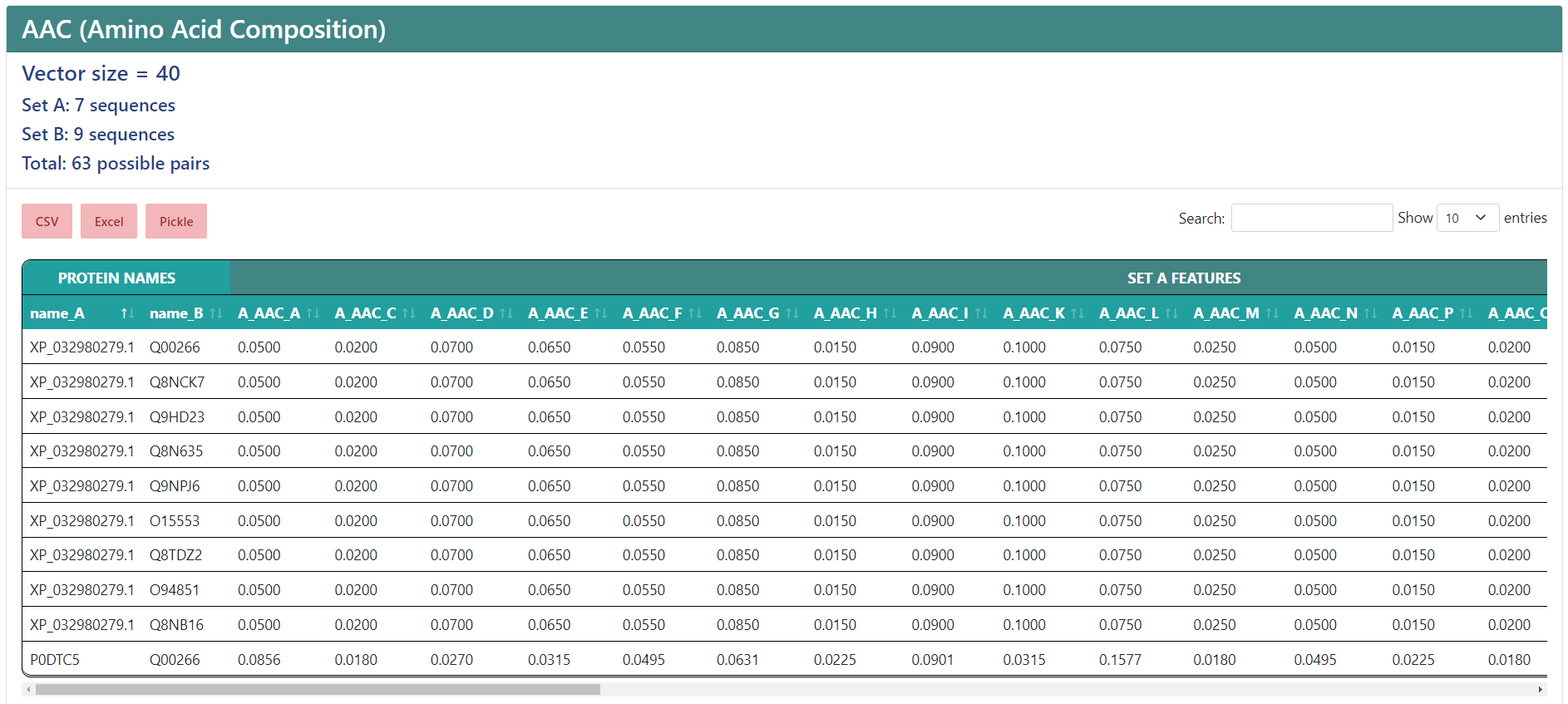
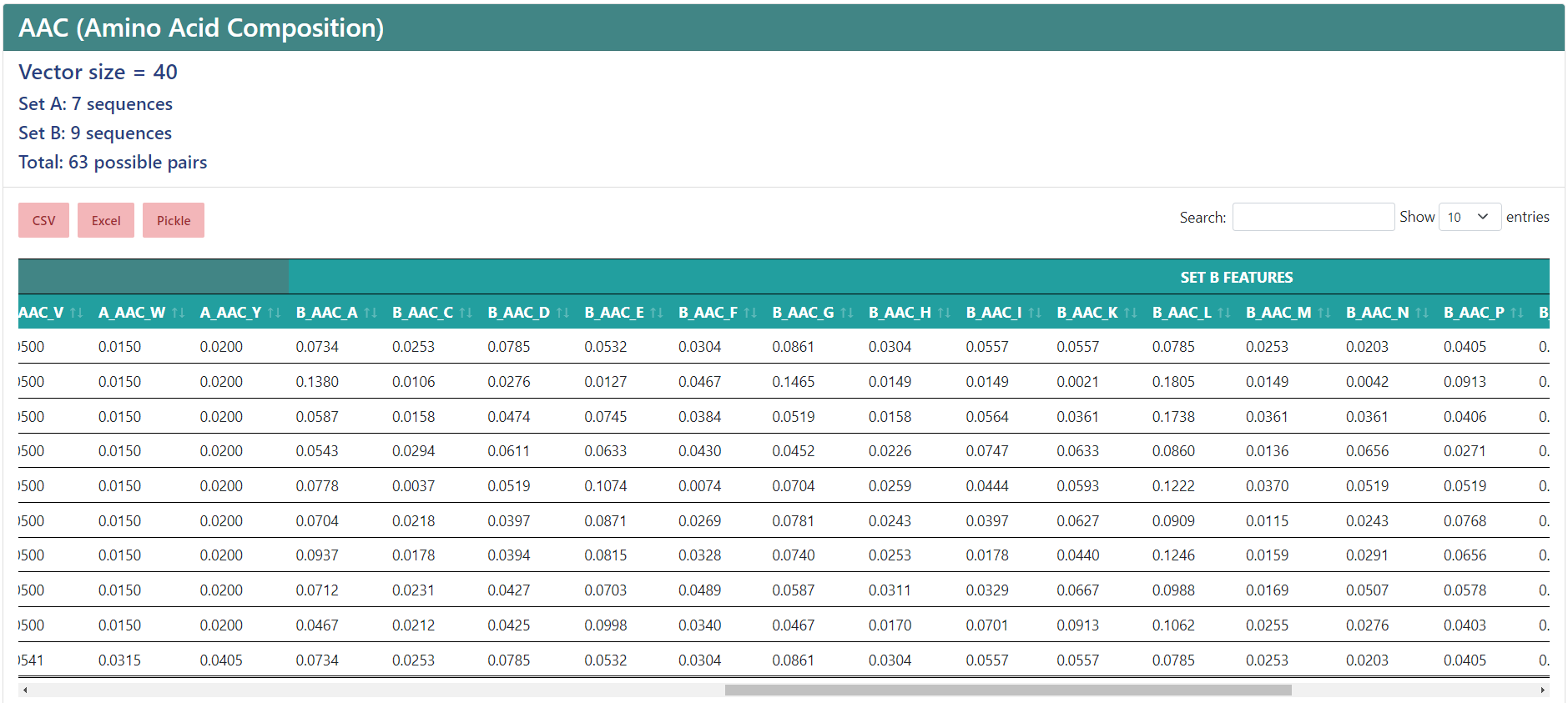
Protein-protein interactions
If you chose to encode protein-protein interactions, under the vector size it will show the number of sequences in the first set (A), the number of sequences in the second set (B) and the total number of interactions. In the results table, the protein names from sets A and B will be separated in two columns, and the feature names will start by "A_" or "B_", depending on whether if sequence the column belongs to is from set A or set B, while having a more general header for those features that belong to the protein from set A and the protein from set B.