Input data and parameters
QualiMap command line
| qualimap bamqc -bam ../mtuberculosis2/2-alignment/pathogen/bam/mycobacteriumTuberculosis/SRR25787977_hisat2.bam -nw 400 -hm 3 |
Alignment
| Command line: | "/home/dguevara/pipeline/src/../tools/hisat2-2.1.0/hisat2-align-s --wrapper basic-0 -x ../mtuberculosis2/2-alignment/pathogen/indices/hisat2/mycobacteriumTuberculosis -S ../mtuberculosis2/2-alignment/pathogen/sam/mycobacteriumTuberculosis/SRR25787977_hisat2.sam -p 16 -1 ../mtuberculosis2/2-alignment/host/fastq/SRR25787977_1.fastq -2 ../mtuberculosis2/2-alignment/host/fastq/SRR25787977_2.fastq" |
| Draw chromosome limits: | no |
| Analyze overlapping paired-end reads: | no |
| Program: | hisat2 (2.1.0) |
| Analysis date: | Thu Feb 01 05:16:23 CST 2024 |
| Size of a homopolymer: | 3 |
| Skip duplicate alignments: | no |
| Number of windows: | 400 |
| BAM file: | ../mtuberculosis2/2-alignment/pathogen/bam/mycobacteriumTuberculosis/SRR25787977_hisat2.bam |
Summary
Globals
| Reference size | 4,411,532 |
| Number of reads | 26,662,258 |
| Mapped reads | 25,652,129 / 96.21% |
| Unmapped reads | 1,010,129 / 3.79% |
| Mapped paired reads | 25,652,129 / 96.21% |
| Mapped reads, first in pair | 12,829,276 / 48.12% |
| Mapped reads, second in pair | 12,822,853 / 48.09% |
| Mapped reads, both in pair | 25,379,585 / 95.19% |
| Mapped reads, singletons | 272,544 / 1.02% |
| Read min/max/mean length | 100 / 100 / 100 |
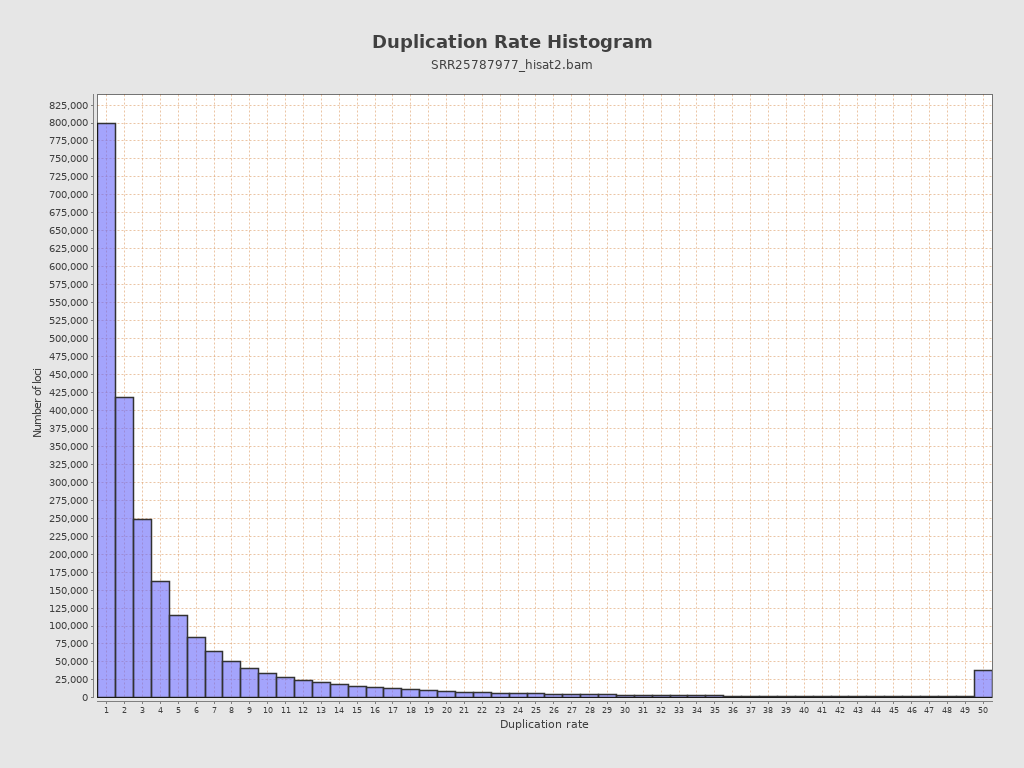
| Duplicated reads (estimated) | 23,335,611 / 87.52% |
| Duplication rate | 65.5% |
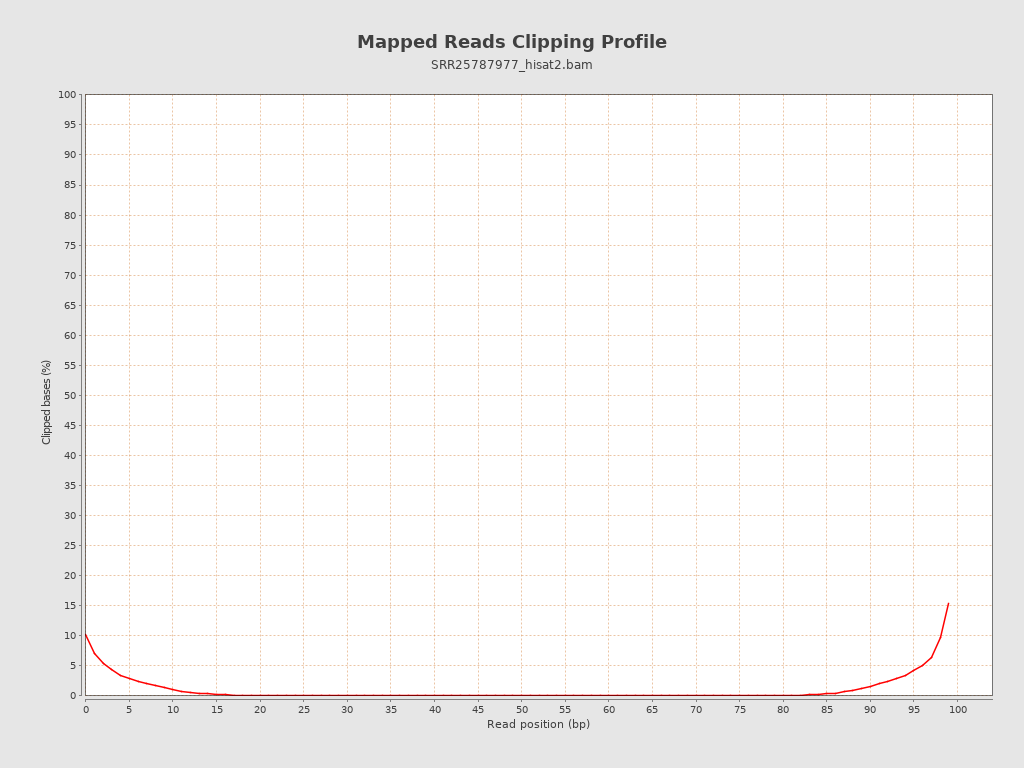
| Clipped reads | 3,059,209 / 11.47% |
ACGT Content
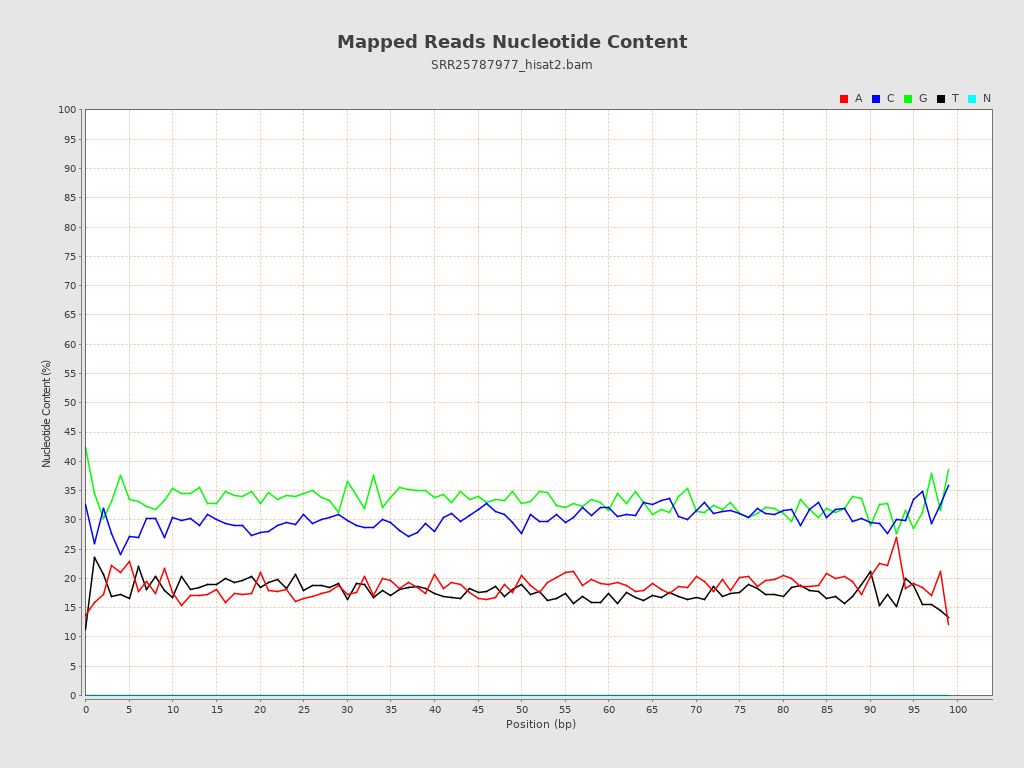
| Number/percentage of A's | 477,884,566 / 18.72% |
| Number/percentage of C's | 772,947,140 / 30.28% |
| Number/percentage of T's | 452,978,233 / 17.74% |
| Number/percentage of G's | 848,992,249 / 33.26% |
| Number/percentage of N's | 253,351,961,114 / 9924.47% |
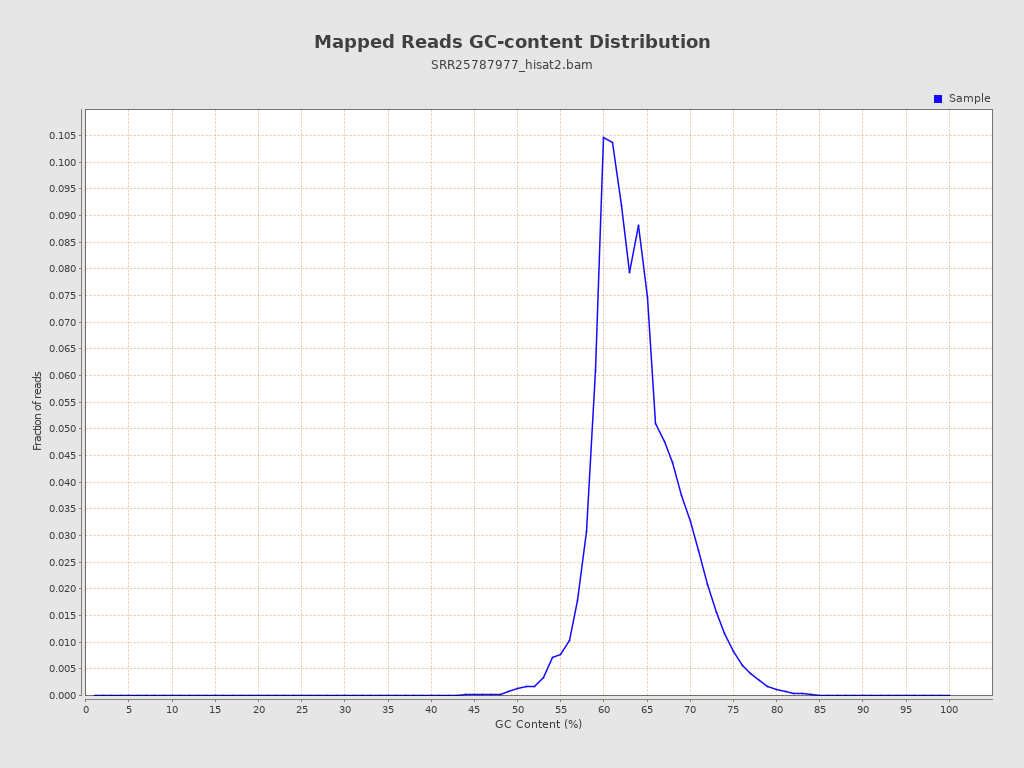
| GC Percentage | 63.54% |
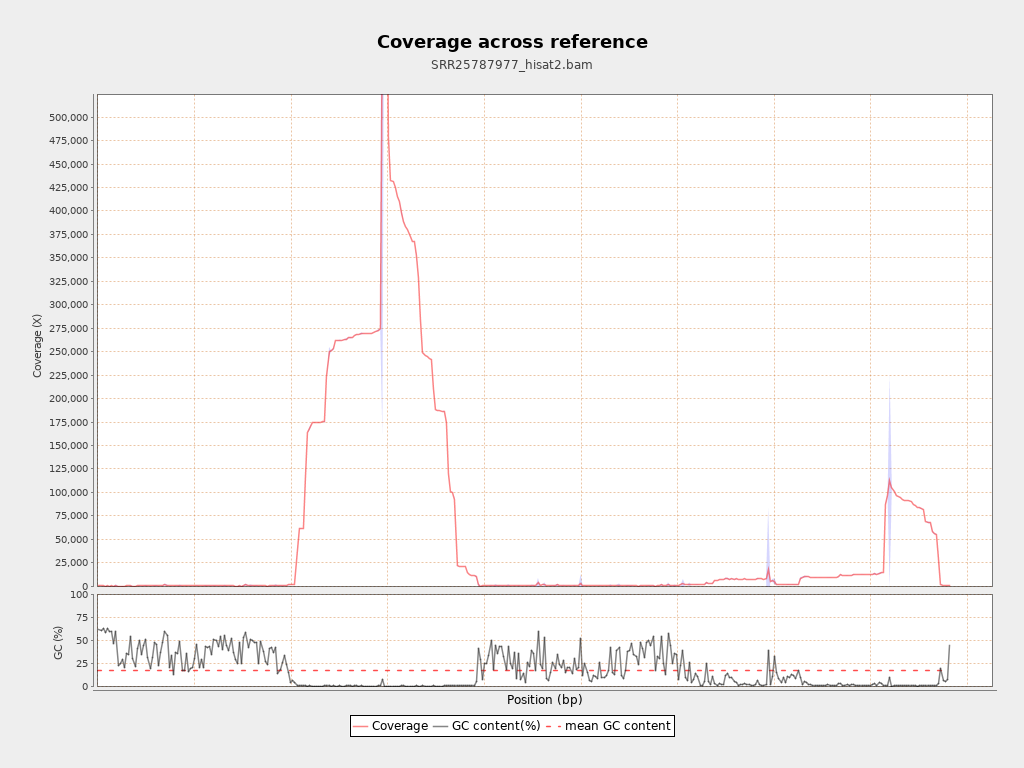
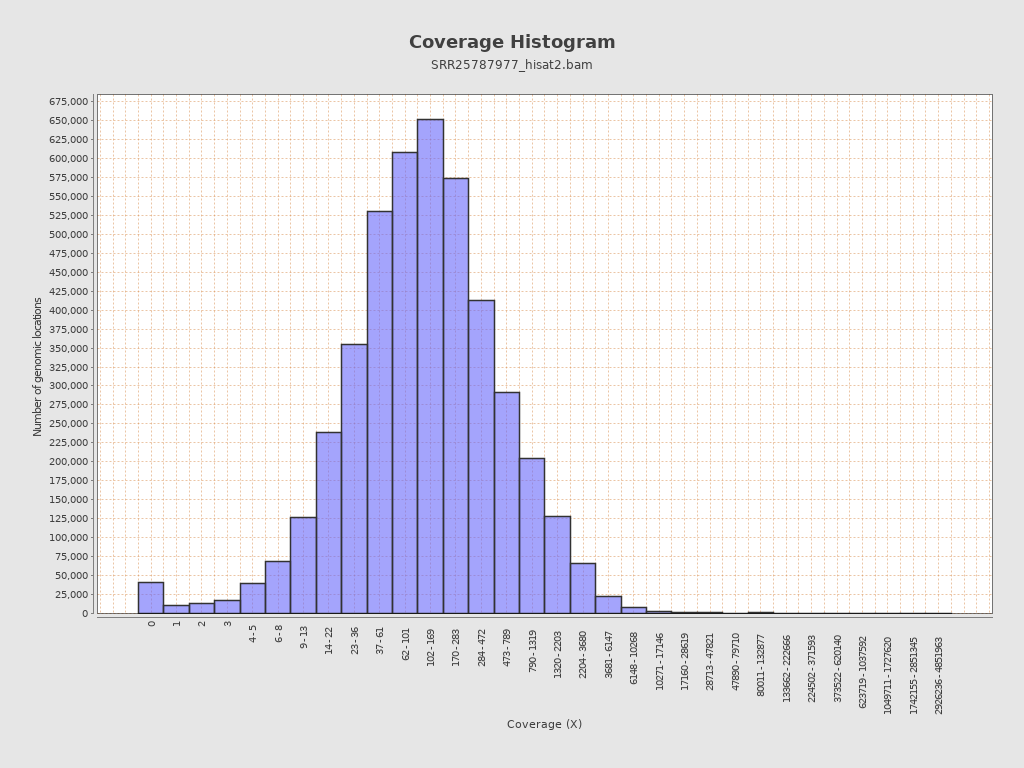
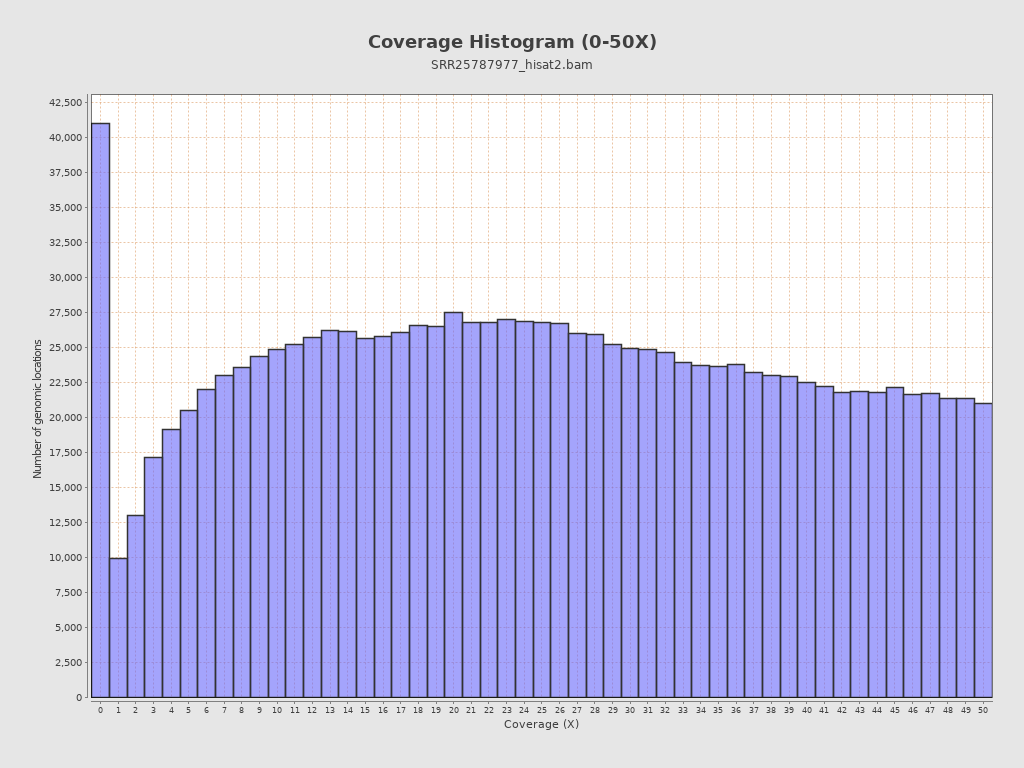
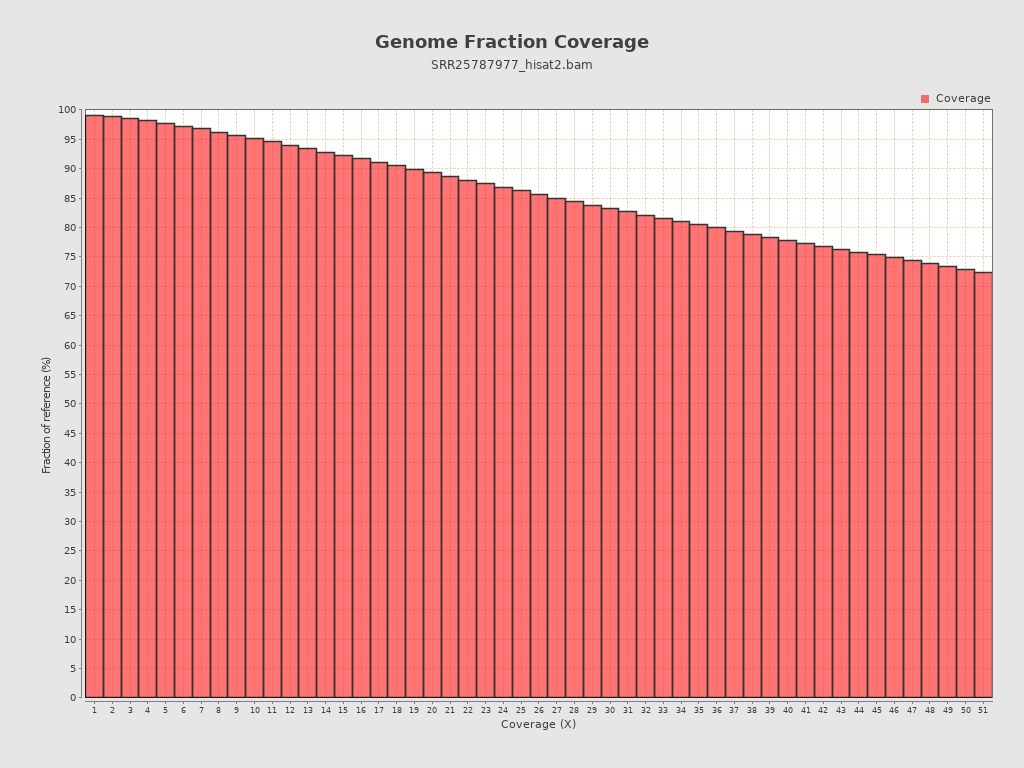
Coverage
| Mean | 58,008.208 |
| Standard Deviation | 62,454.0823 |
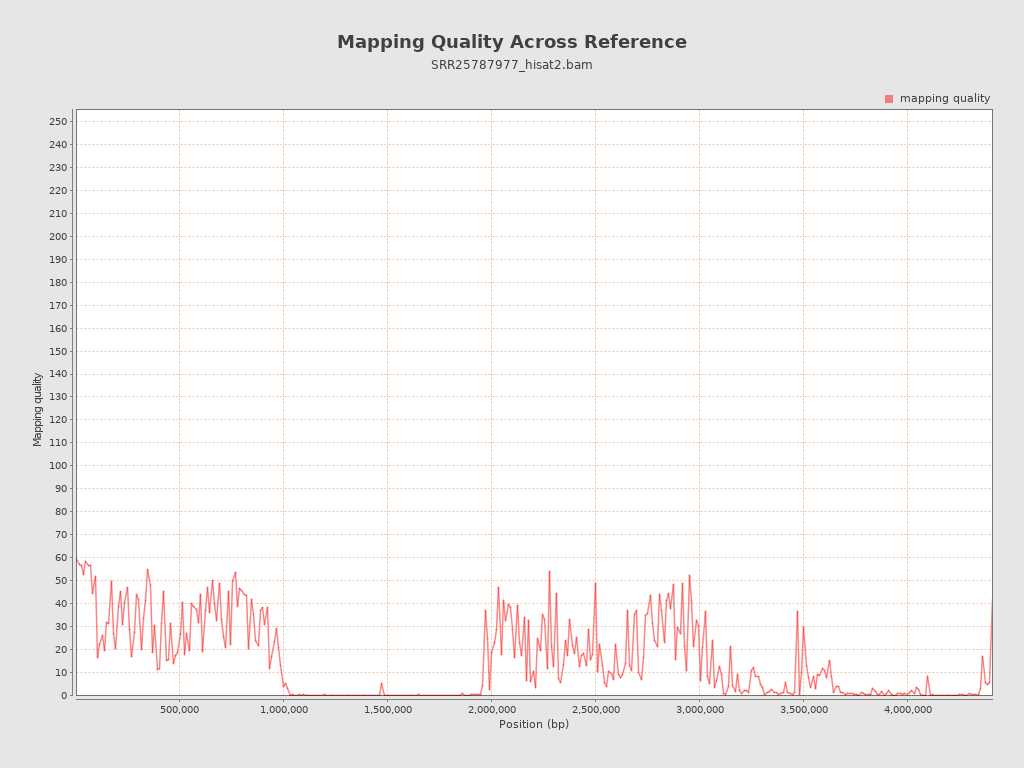
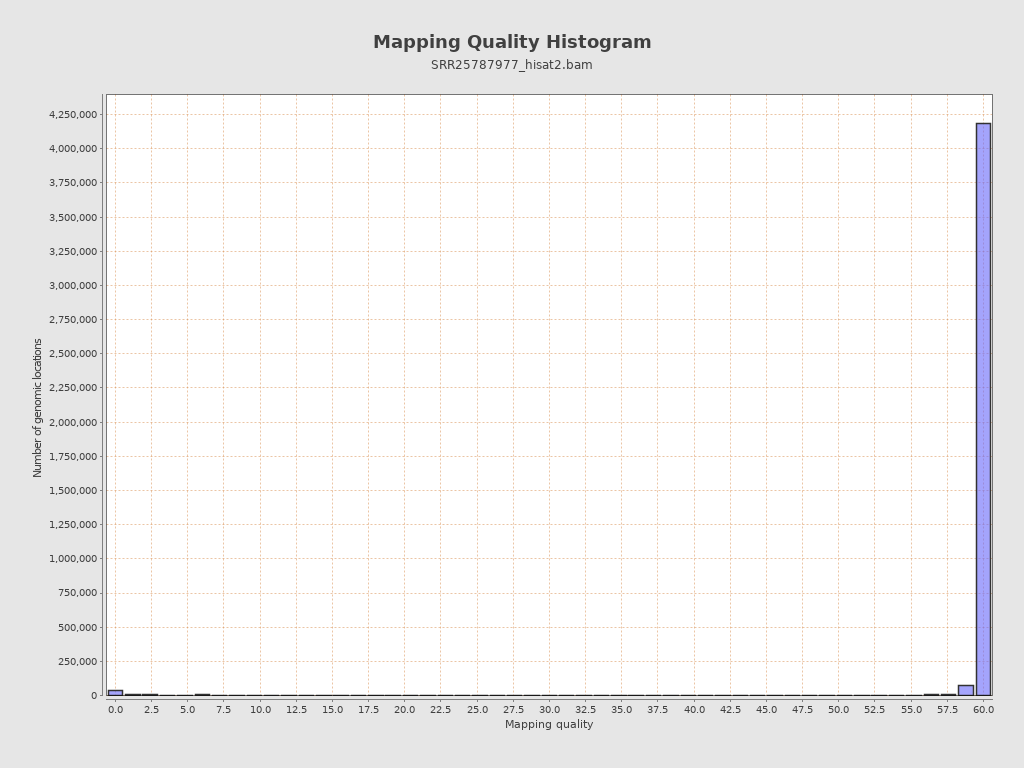
Mapping Quality
| Mean Mapping Quality | 14.87 |
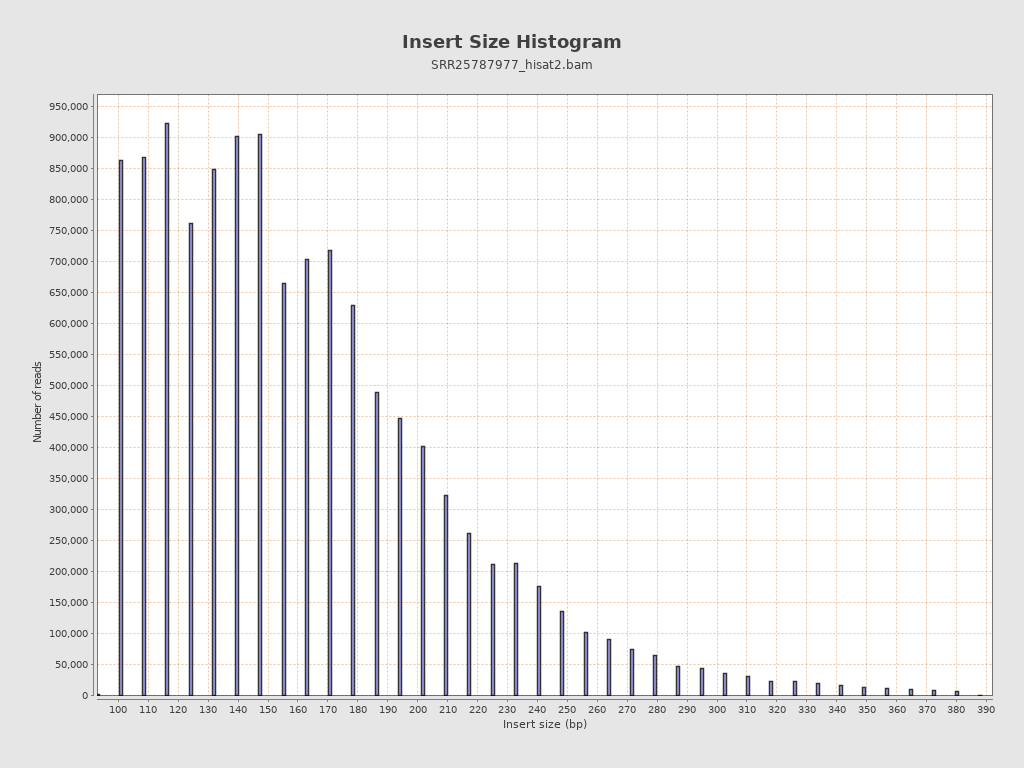
Insert size
| Mean | 26,929.44 |
| Standard Deviation | 238,024.67 |
| P25/Median/P75 | 129 / 157 / 194 |
Mismatches and indels
| General error rate | 0% |
| Mismatches | 5,442,455 |
| Insertions | 10,843 |
| Mapped reads with at least one insertion | 0.04% |
| Deletions | 238,863 |
| Mapped reads with at least one deletion | 0.93% |
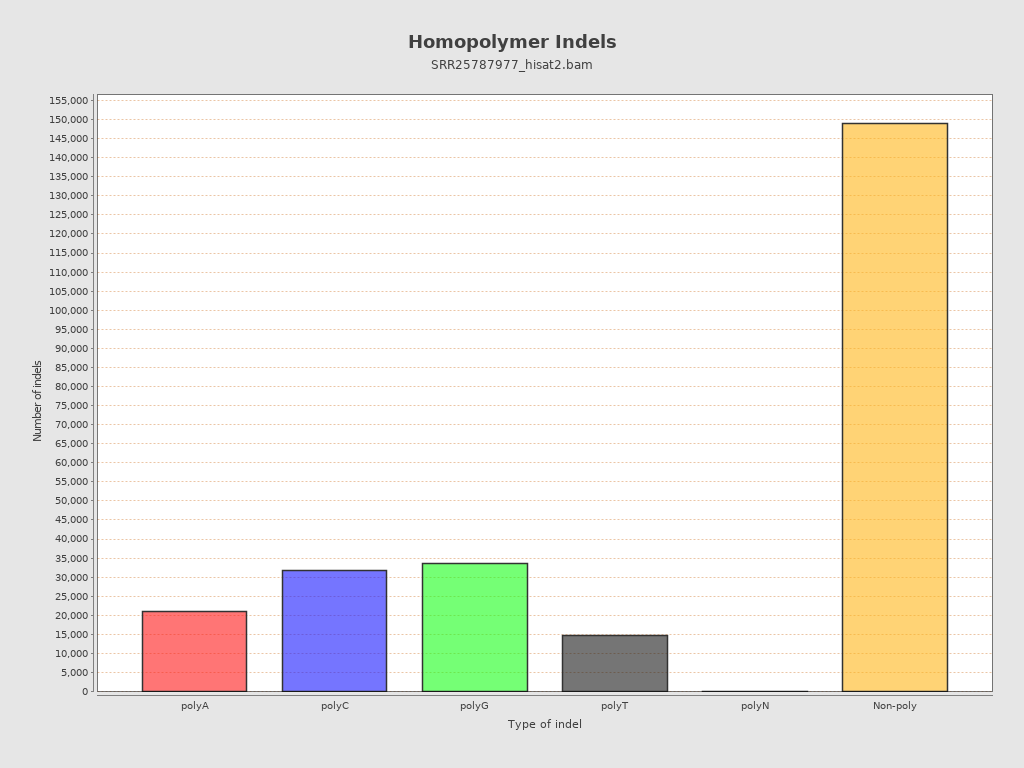
| Homopolymer indels | 40.33% |
Chromosome stats
| Name | Length | Mapped bases | Mean coverage | Standard deviation |
| AL123456.3 | 4411532 | 255905065705 | 58,008.208 | 62,454.0823 |