Input data and parameters
QualiMap command line
| qualimap bamqc -bam ../mtuberculosis2/2-alignment/pathogen/bam/mycobacteriumTuberculosis/SRR25787973_hisat2.bam -nw 400 -hm 3 |
Alignment
| Command line: | "/home/dguevara/pipeline/src/../tools/hisat2-2.1.0/hisat2-align-s --wrapper basic-0 -x ../mtuberculosis2/2-alignment/pathogen/indices/hisat2/mycobacteriumTuberculosis -S ../mtuberculosis2/2-alignment/pathogen/sam/mycobacteriumTuberculosis/SRR25787973_hisat2.sam -p 16 -1 ../mtuberculosis2/2-alignment/host/fastq/SRR25787973_1.fastq -2 ../mtuberculosis2/2-alignment/host/fastq/SRR25787973_2.fastq" |
| Draw chromosome limits: | no |
| Analyze overlapping paired-end reads: | no |
| Program: | hisat2 (2.1.0) |
| Analysis date: | Thu Feb 01 04:43:50 CST 2024 |
| Size of a homopolymer: | 3 |
| Skip duplicate alignments: | no |
| Number of windows: | 400 |
| BAM file: | ../mtuberculosis2/2-alignment/pathogen/bam/mycobacteriumTuberculosis/SRR25787973_hisat2.bam |
Summary
Globals
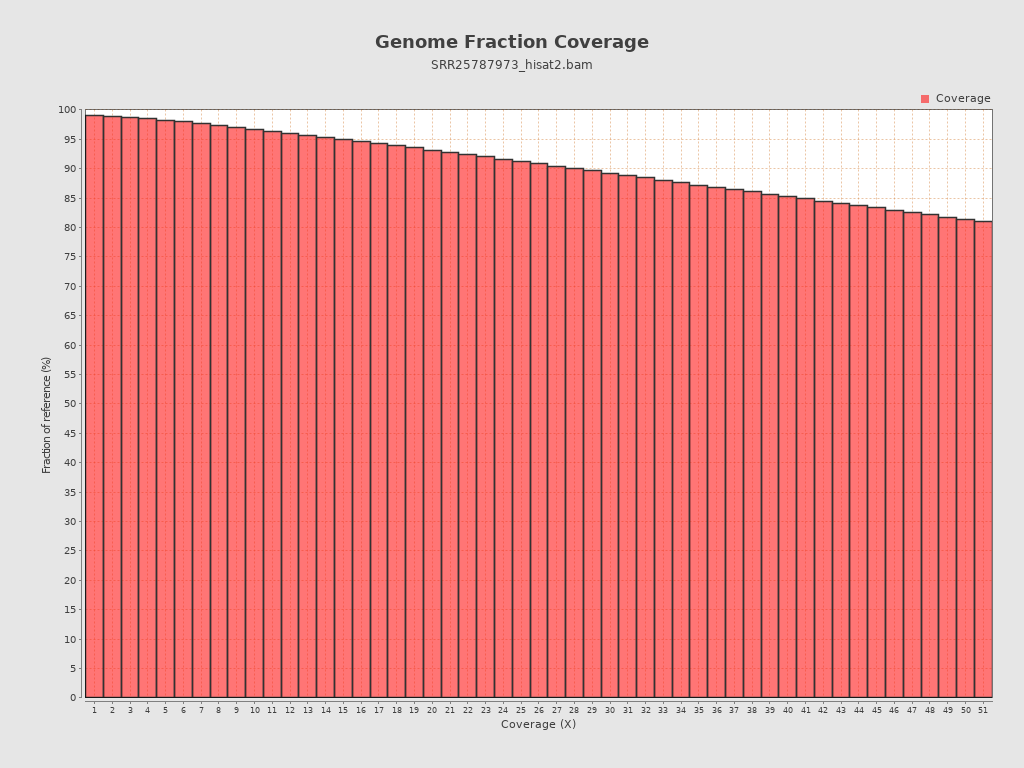
| Reference size | 4,411,532 |
| Number of reads | 30,696,554 |
| Mapped reads | 29,624,550 / 96.51% |
| Unmapped reads | 1,072,004 / 3.49% |
| Mapped paired reads | 29,624,550 / 96.51% |
| Mapped reads, first in pair | 14,802,984 / 48.22% |
| Mapped reads, second in pair | 14,821,566 / 48.28% |
| Mapped reads, both in pair | 29,408,785 / 95.8% |
| Mapped reads, singletons | 215,765 / 0.7% |
| Read min/max/mean length | 100 / 100 / 100 |
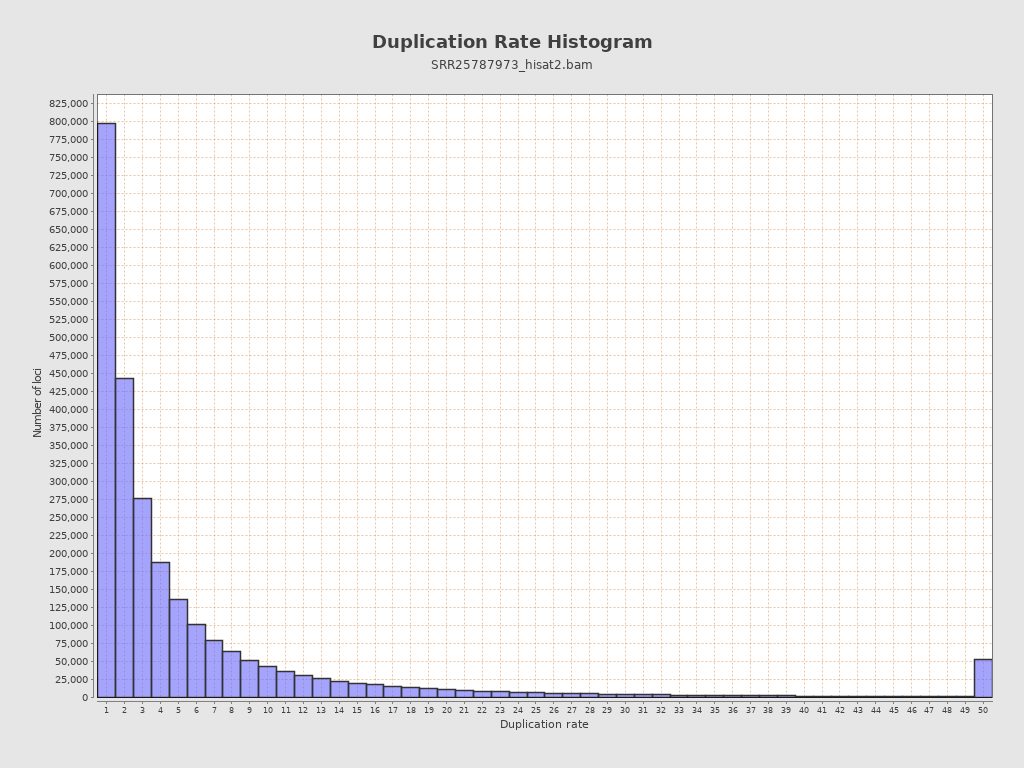
| Duplicated reads (estimated) | 27,054,757 / 88.14% |
| Duplication rate | 68.99% |
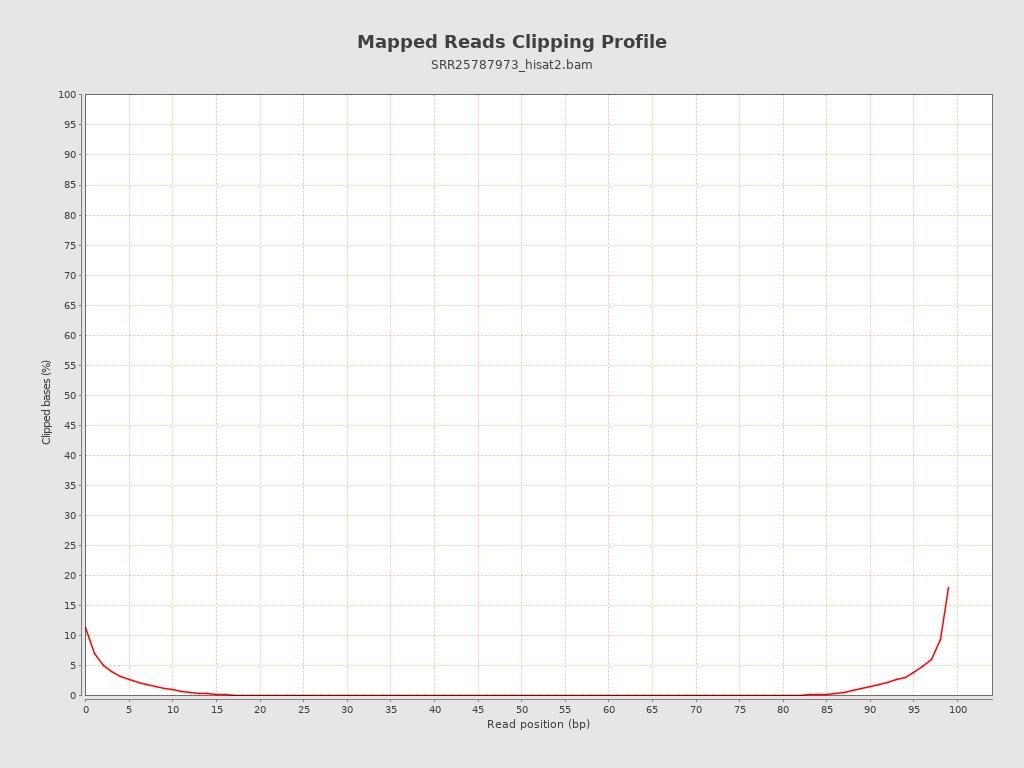
| Clipped reads | 2,560,387 / 8.34% |
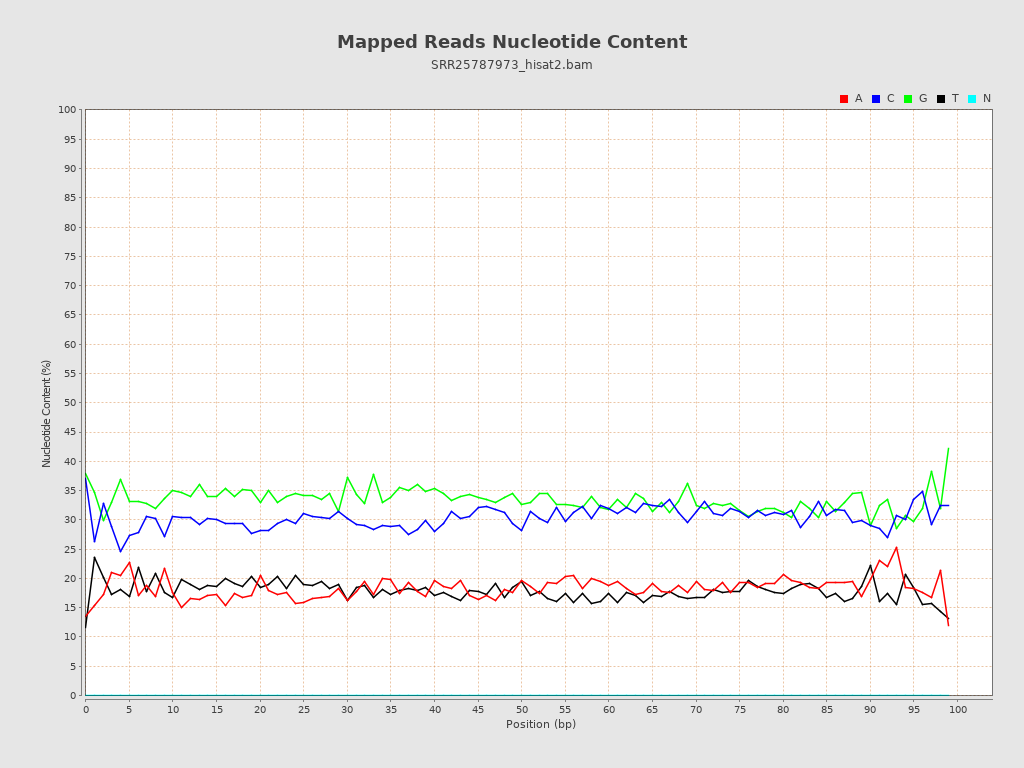
ACGT Content
| Number/percentage of A's | 541,524,536 / 18.34% |
| Number/percentage of C's | 897,672,803 / 30.39% |
| Number/percentage of T's | 526,554,250 / 17.83% |
| Number/percentage of G's | 987,744,728 / 33.44% |
| Number/percentage of N's | 112,789,006,767 / 3818.83% |
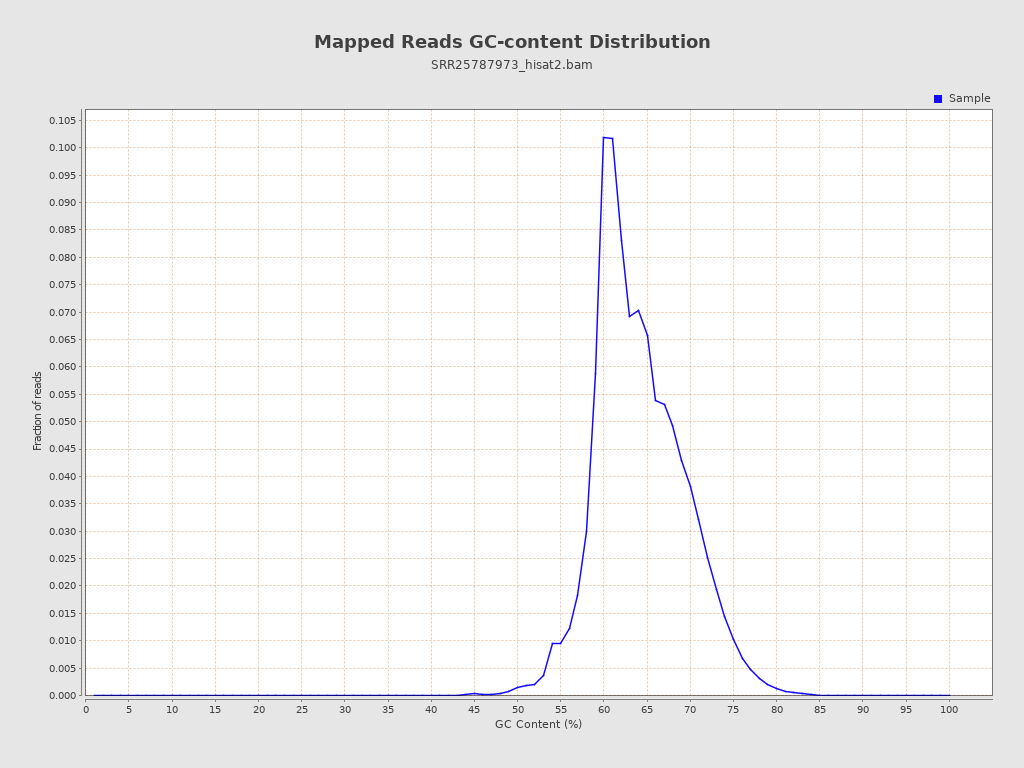
| GC Percentage | 63.84% |
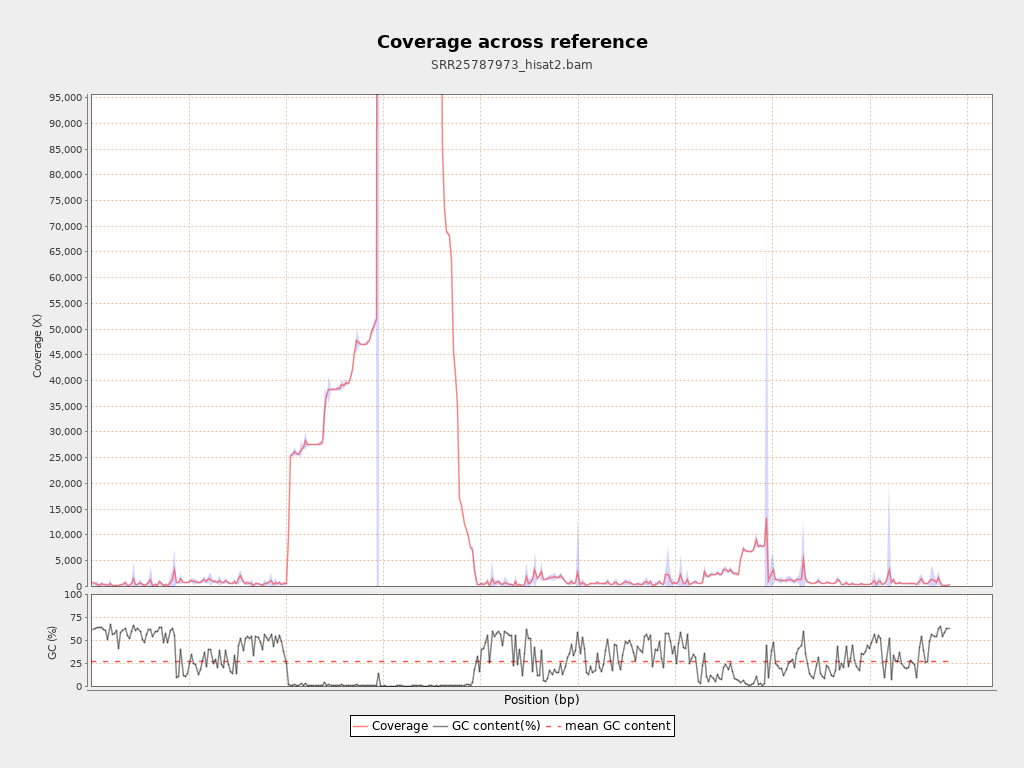
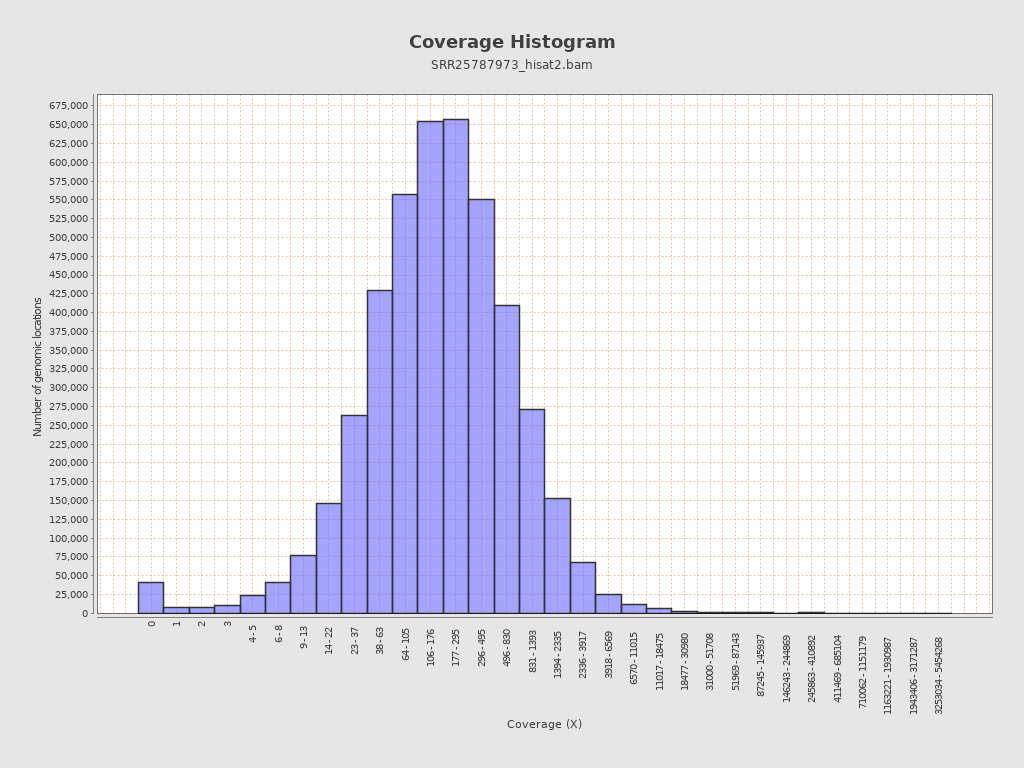
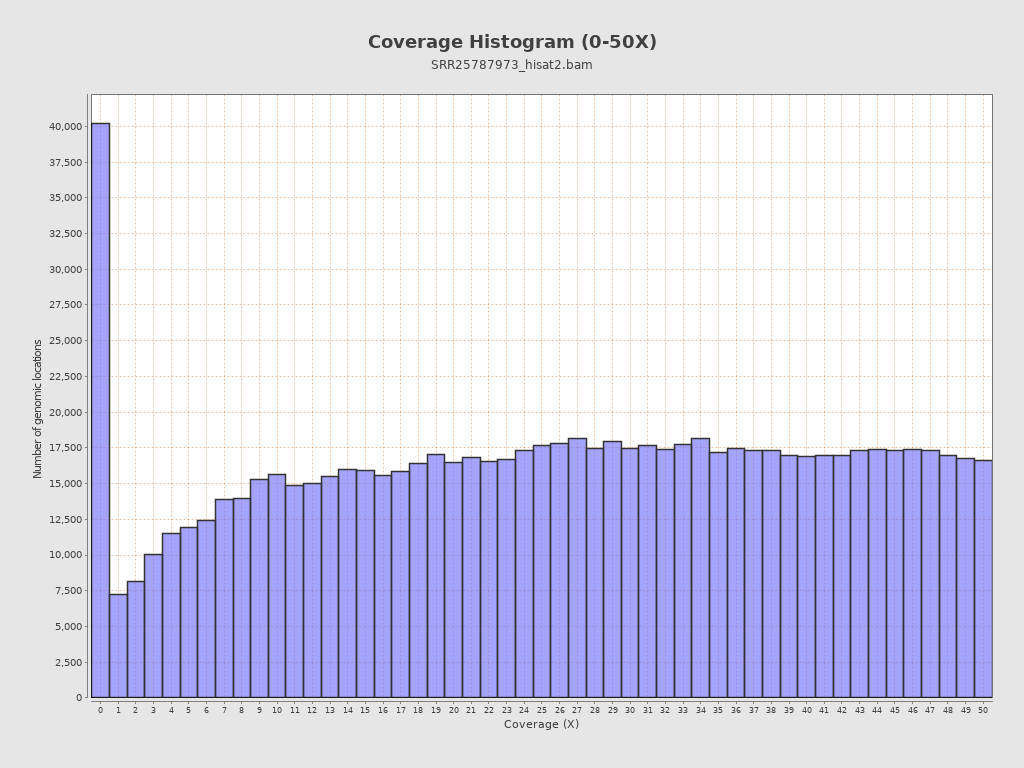
Coverage
| Mean | 26,236.373 |
| Standard Deviation | 36,334.7785 |
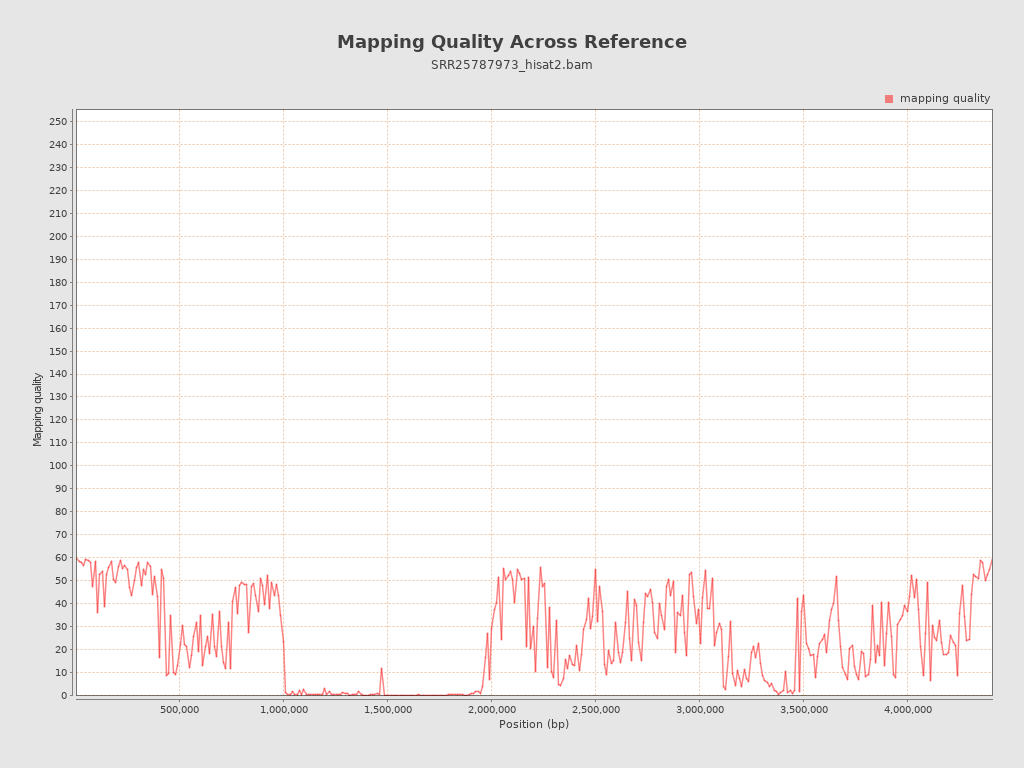
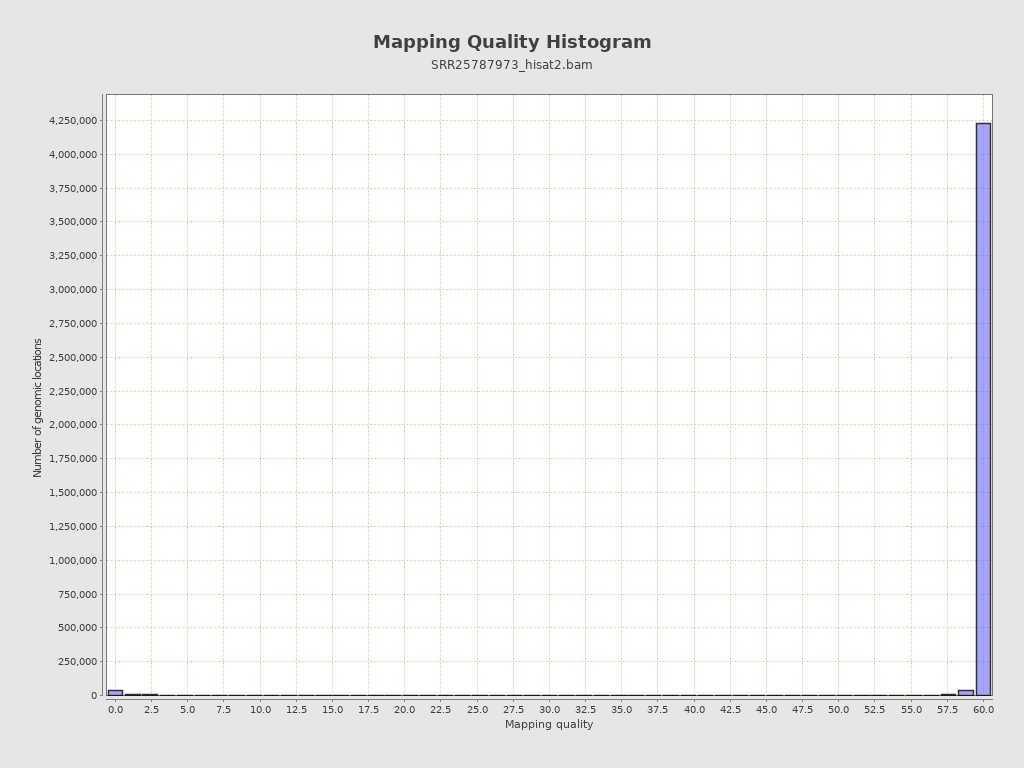
Mapping Quality
| Mean Mapping Quality | 24.35 |
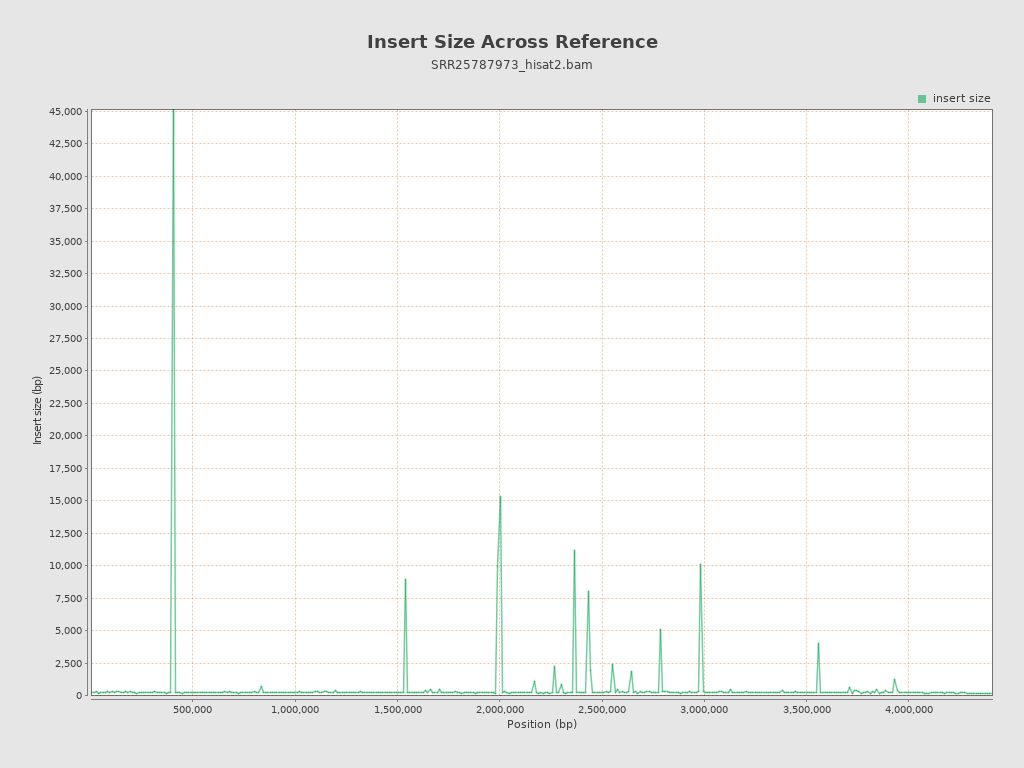
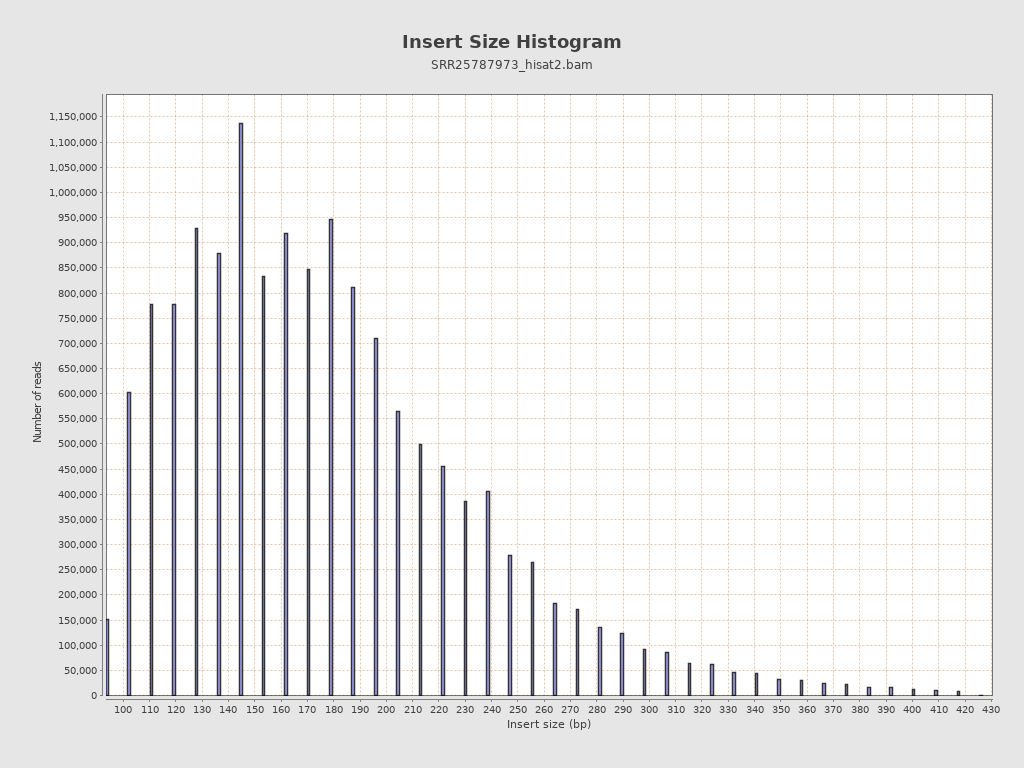
Insert size
| Mean | 8,821.01 |
| Standard Deviation | 131,789.52 |
| P25/Median/P75 | 140 / 173 / 213 |
Mismatches and indels
| General error rate | 0% |
| Mismatches | 3,565,057 |
| Insertions | 16,139 |
| Mapped reads with at least one insertion | 0.05% |
| Deletions | 68,832 |
| Mapped reads with at least one deletion | 0.23% |
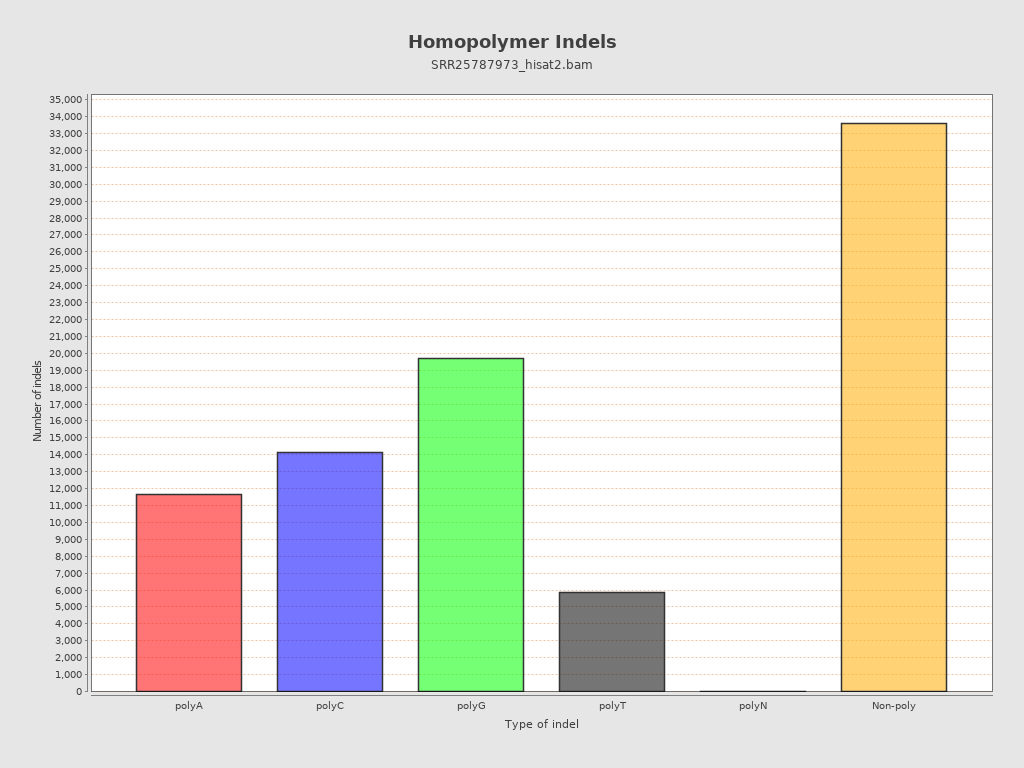
| Homopolymer indels | 60.45% |
Chromosome stats
| Name | Length | Mapped bases | Mean coverage | Standard deviation |
| AL123456.3 | 4411532 | 115742599173 | 26,236.373 | 36,334.7785 |