Input data and parameters
QualiMap command line
| qualimap bamqc -bam ../influenzaA/2-alignment/pathogen/bam/influenzaA/DRR051450_bwa.bam -nw 400 -hm 3 |
Alignment
| Command line: | bwa mem -t 16 ../influenzaA/2-alignment/pathogen/indices/bwa/influenzaA ../influenzaA/2-alignment/host/fastq/DRR051450_1.fastq ../influenzaA/2-alignment/host/fastq/DRR051450_2.fastq |
| Draw chromosome limits: | no |
| Analyze overlapping paired-end reads: | no |
| Program: | bwa (0.7.17-r1188) |
| Analysis date: | Wed Jan 17 22:08:18 CST 2024 |
| Size of a homopolymer: | 3 |
| Skip duplicate alignments: | no |
| Number of windows: | 400 |
| BAM file: | ../influenzaA/2-alignment/pathogen/bam/influenzaA/DRR051450_bwa.bam |
Summary
Globals
| Reference size | 13,627 |
| Number of reads | 566,073 |
| Mapped reads | 12,813 / 2.26% |
| Supplementary alignments | 37 / 0.01% |
| Unmapped reads | 553,260 / 97.74% |
| Mapped paired reads | 12,813 / 2.26% |
| Mapped reads, first in pair | 6,402 / 1.13% |
| Mapped reads, second in pair | 6,411 / 1.13% |
| Mapped reads, both in pair | 12,761 / 2.25% |
| Mapped reads, singletons | 52 / 0.01% |
| Read min/max/mean length | 30 / 100 / 100 |
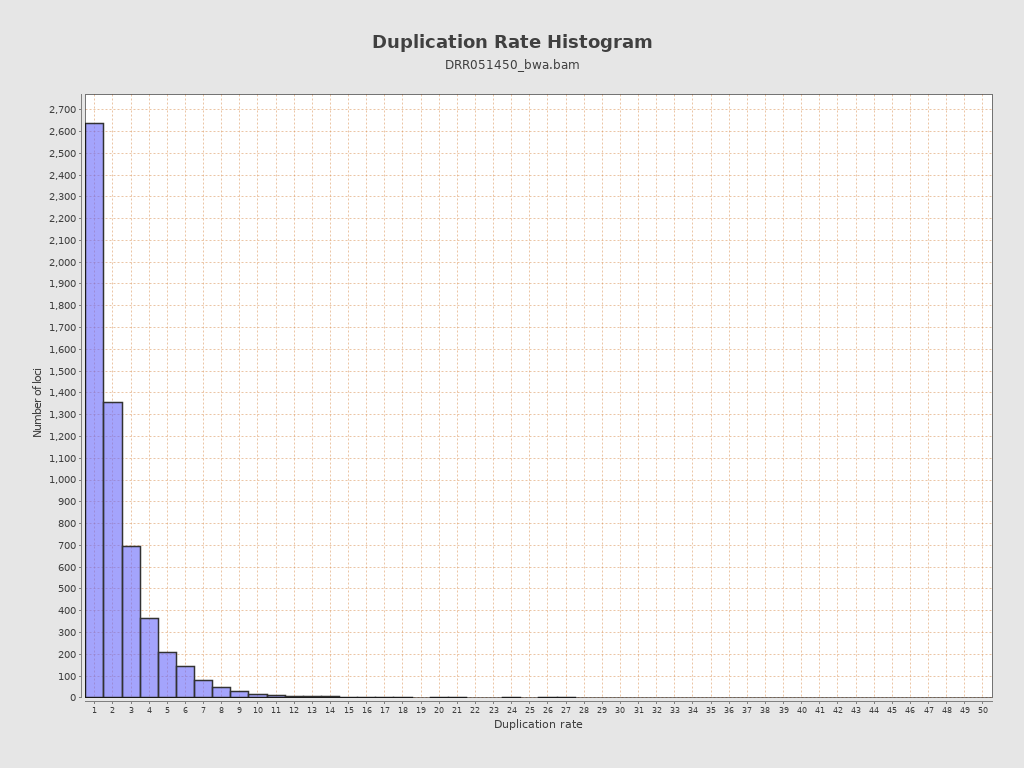
| Duplicated reads (estimated) | 7,191 / 1.27% |
| Duplication rate | 53.11% |
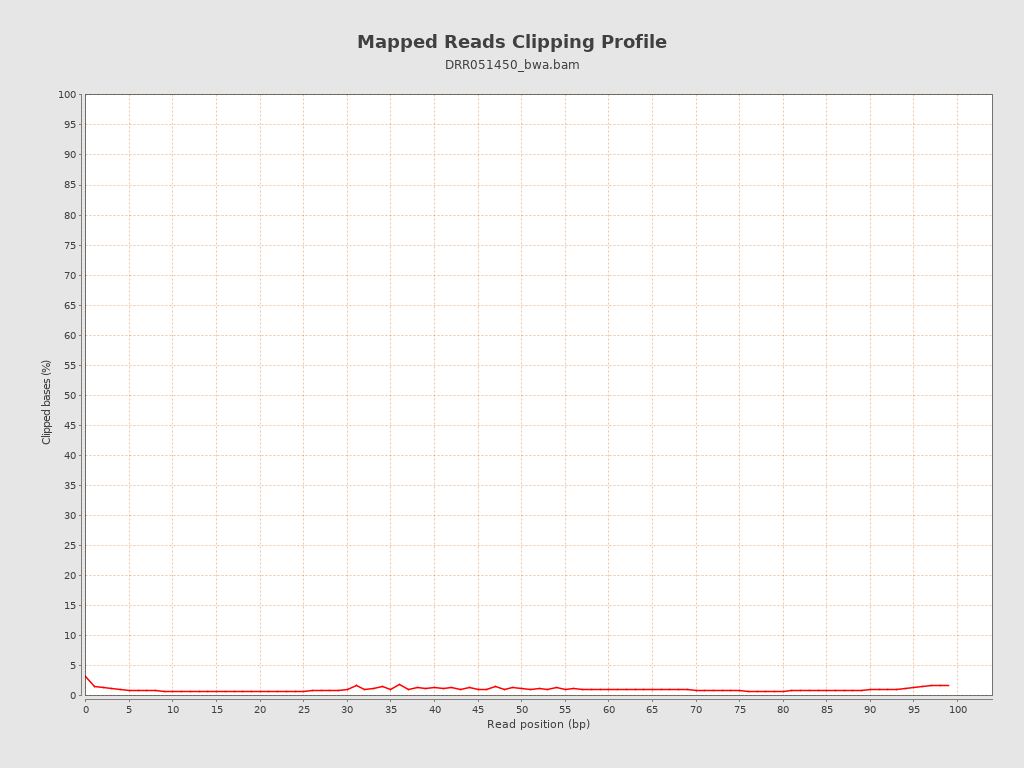
| Clipped reads | 903 / 0.16% |
ACGT Content
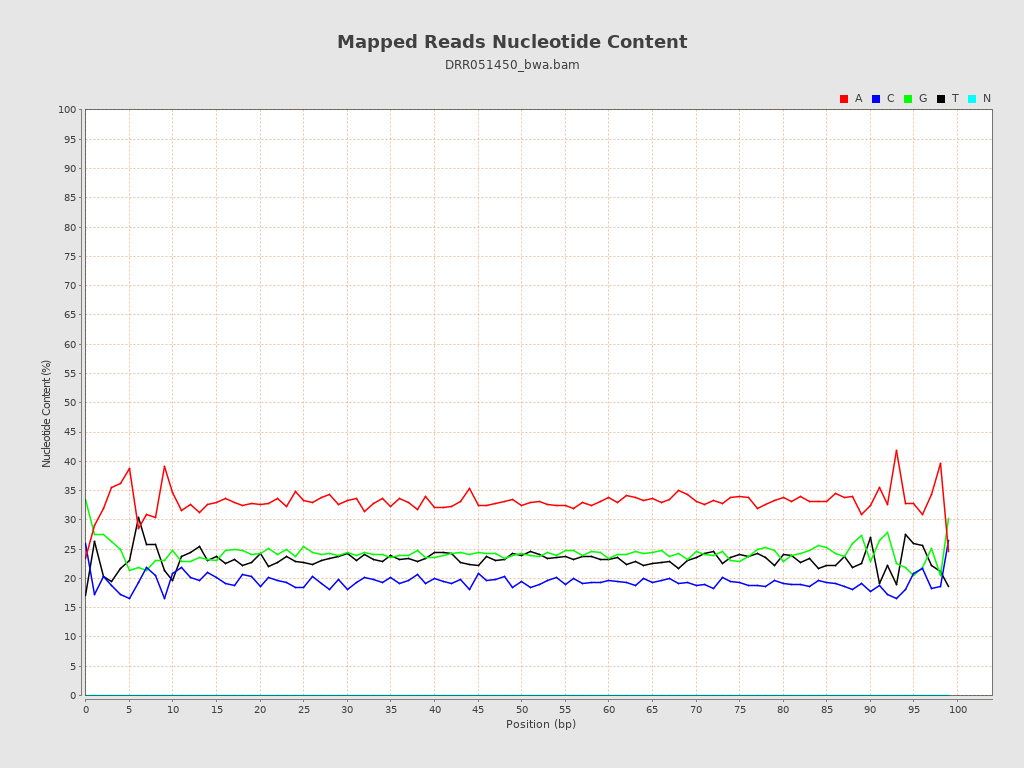
| Number/percentage of A's | 414,797 / 33.1% |
| Number/percentage of C's | 243,573 / 19.43% |
| Number/percentage of T's | 290,641 / 23.19% |
| Number/percentage of G's | 304,331 / 24.28% |
| Number/percentage of N's | 0 / 0% |
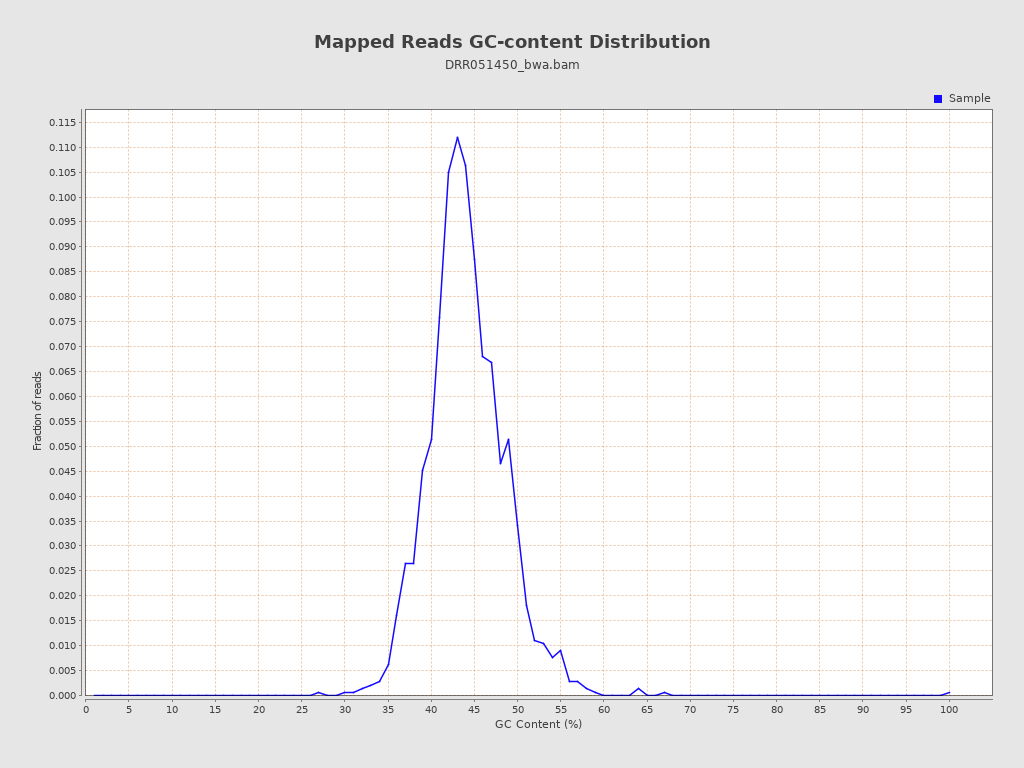
| GC Percentage | 43.72% |
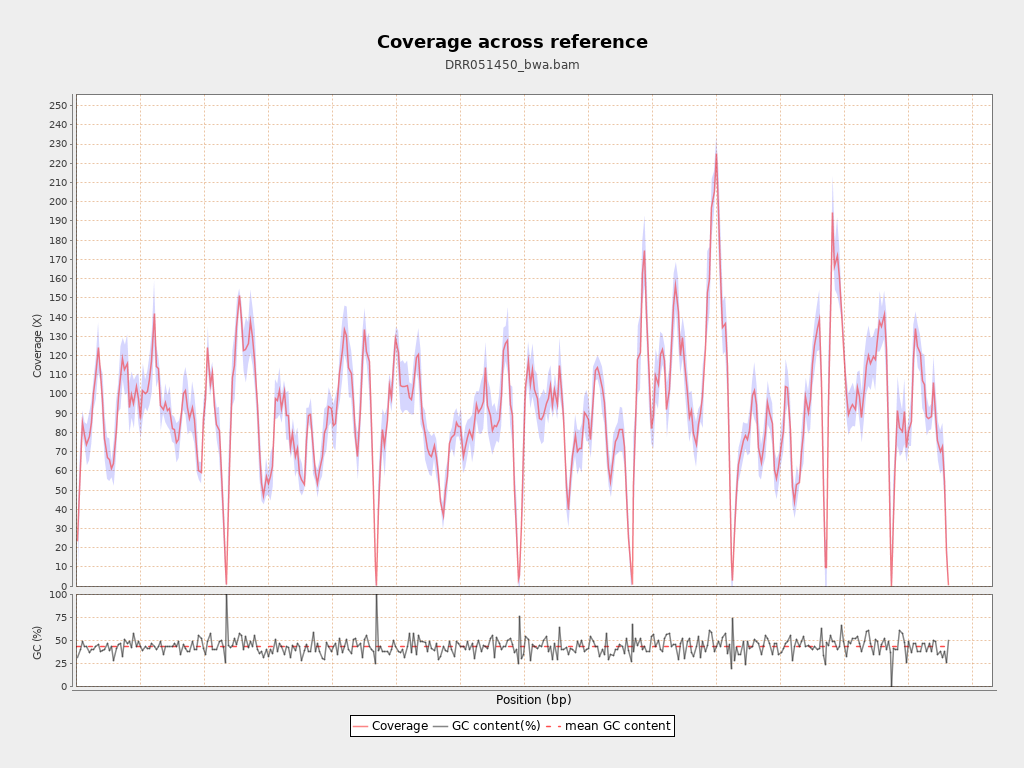
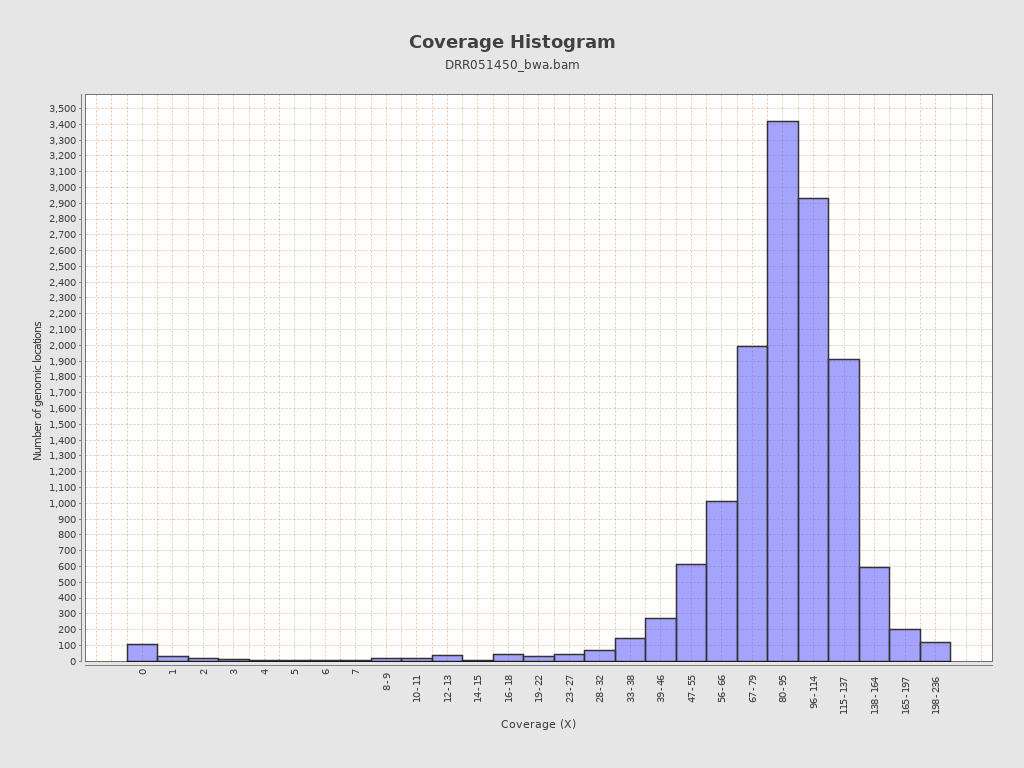
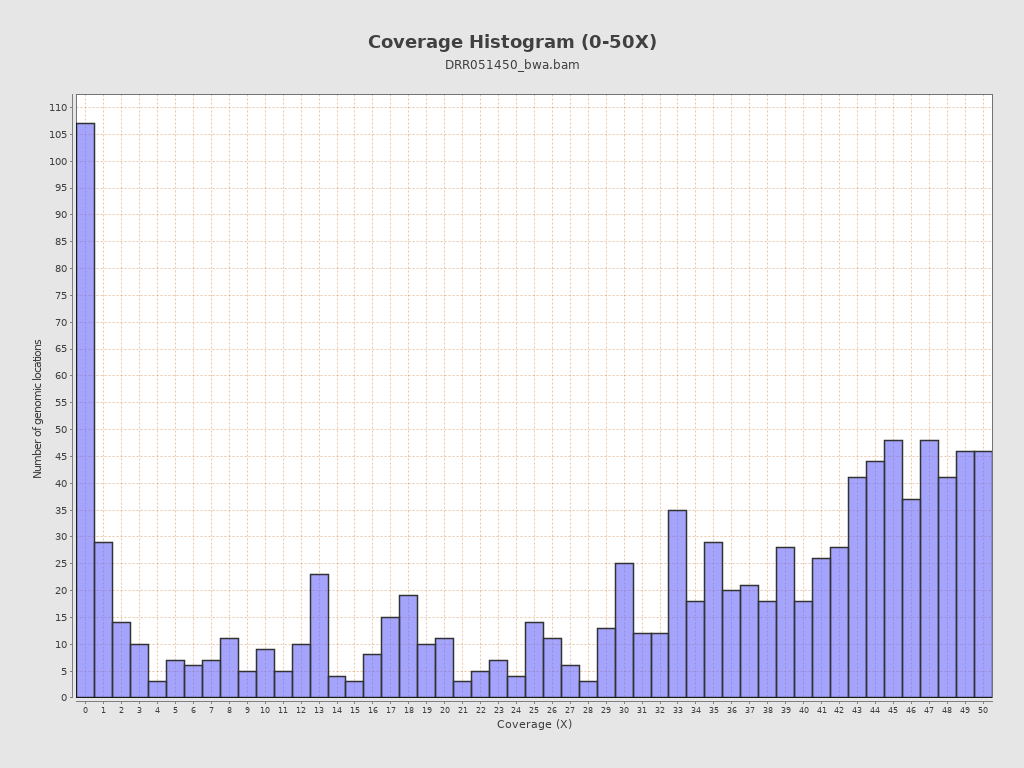
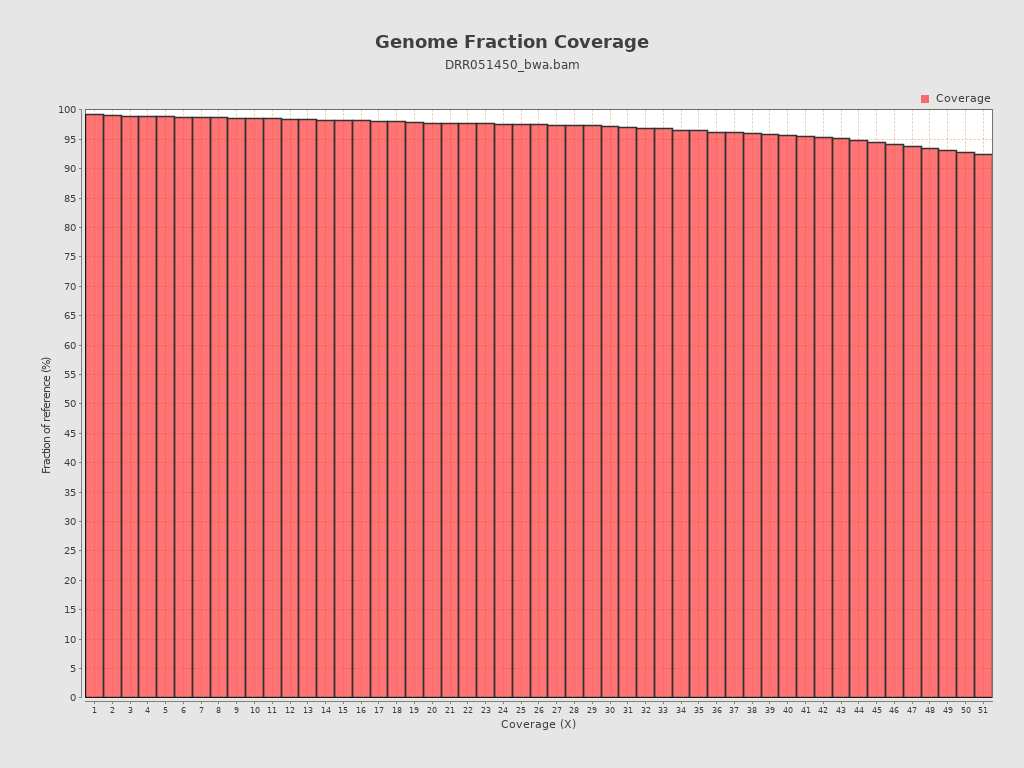
Coverage
| Mean | 91.9892 |
| Standard Deviation | 32.43 |
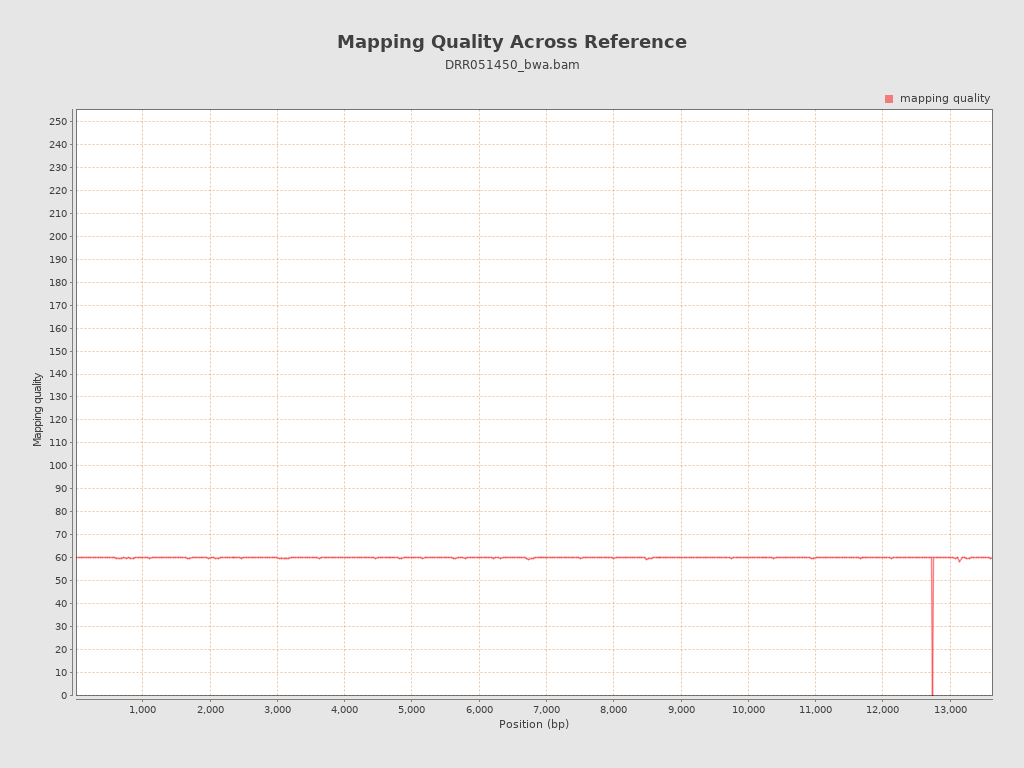
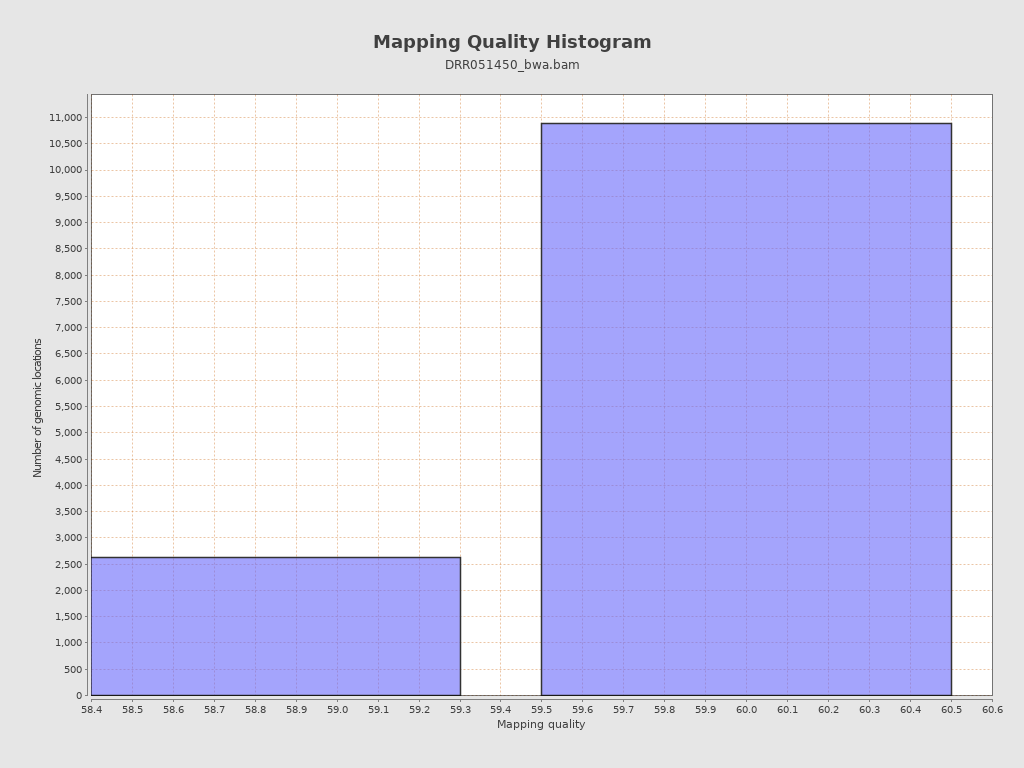
Mapping Quality
| Mean Mapping Quality | 59.81 |
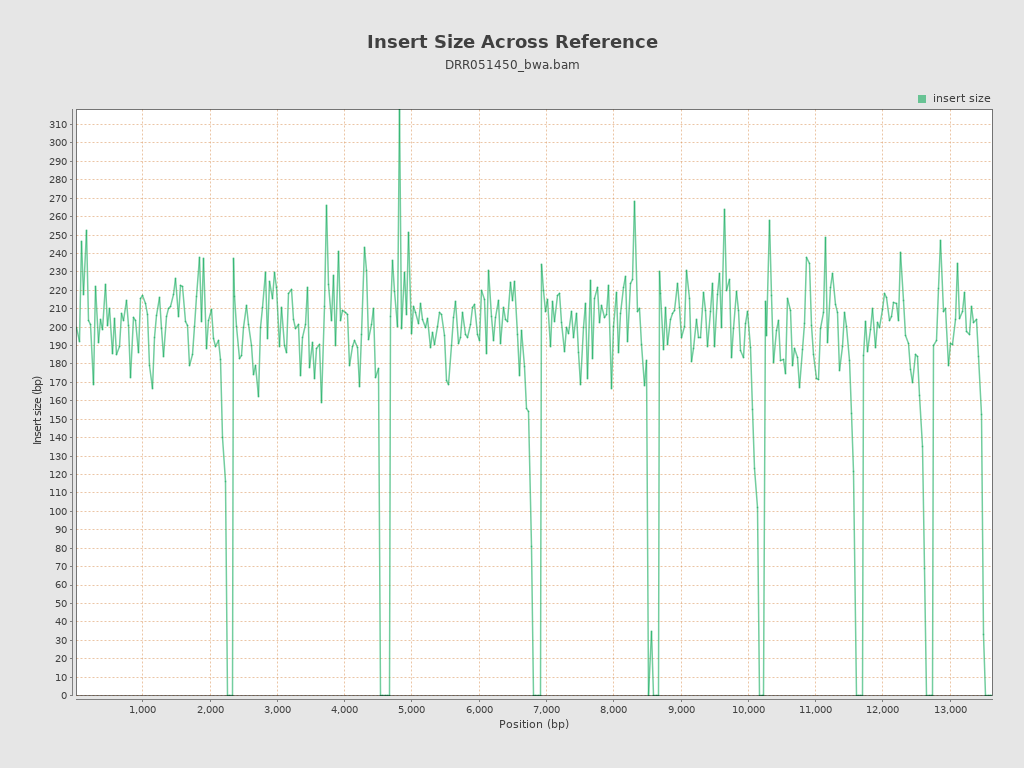
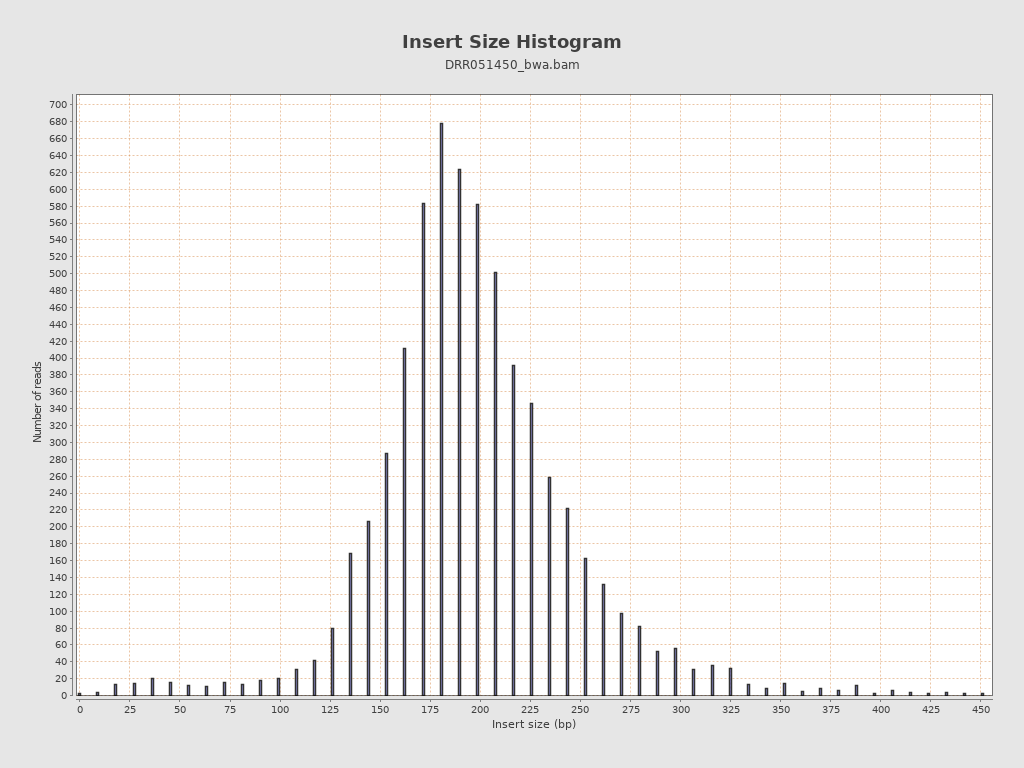
Insert size
| Mean | 206.51 |
| Standard Deviation | 94.92 |
| P25/Median/P75 | 175 / 197 / 227 |
Mismatches and indels
| General error rate | 2.9% |
| Mismatches | 36,333 |
| Insertions | 43 |
| Mapped reads with at least one insertion | 0.34% |
| Deletions | 69 |
| Mapped reads with at least one deletion | 0.54% |
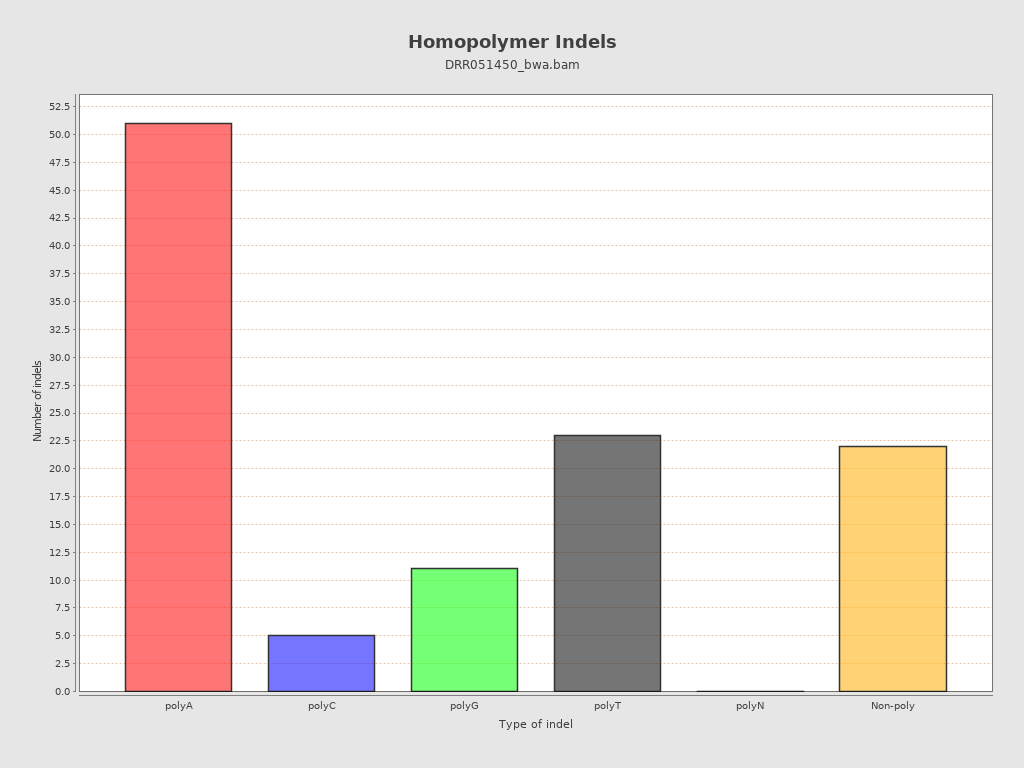
| Homopolymer indels | 80.36% |
Chromosome stats
| Name | Length | Mapped bases | Mean coverage | Standard deviation |
| NC_007373.1 | 2341 | 209298 | 89.4054 | 23.0481 |
| NC_007372.1 | 2341 | 210430 | 89.8889 | 29.711 |
| NC_007371.1 | 2233 | 189553 | 84.8871 | 25.0619 |
| NC_007366.1 | 1762 | 143794 | 81.6084 | 25.7393 |
| NC_007369.1 | 1566 | 189904 | 121.2669 | 41.5392 |
| NC_007368.1 | 1467 | 115815 | 78.9468 | 27.1236 |
| NC_007367.1 | 1027 | 119540 | 116.3973 | 36.1803 |
| NC_007370.1 | 890 | 75203 | 84.4978 | 28.8669 |